biorxiv.org/content/10.110…
It has really open up my eyes to the potential of #computationalbiology, and is the reason why we have a new #datascience position available:
babrahaminstitute.livevacancies.co.uk/#/job/details/…

@CarlyEWhyte can walk you through the whole process in <2'
biorxiv.org/content/10.110…
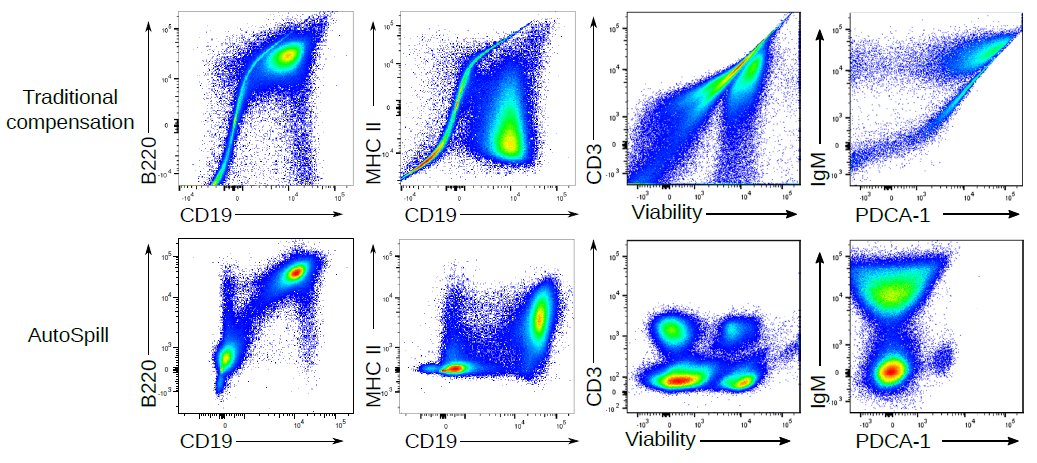
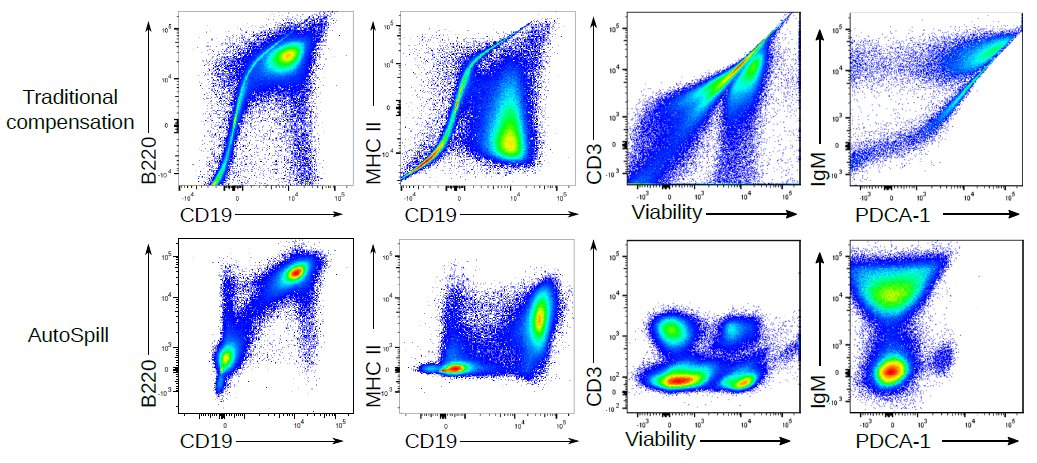
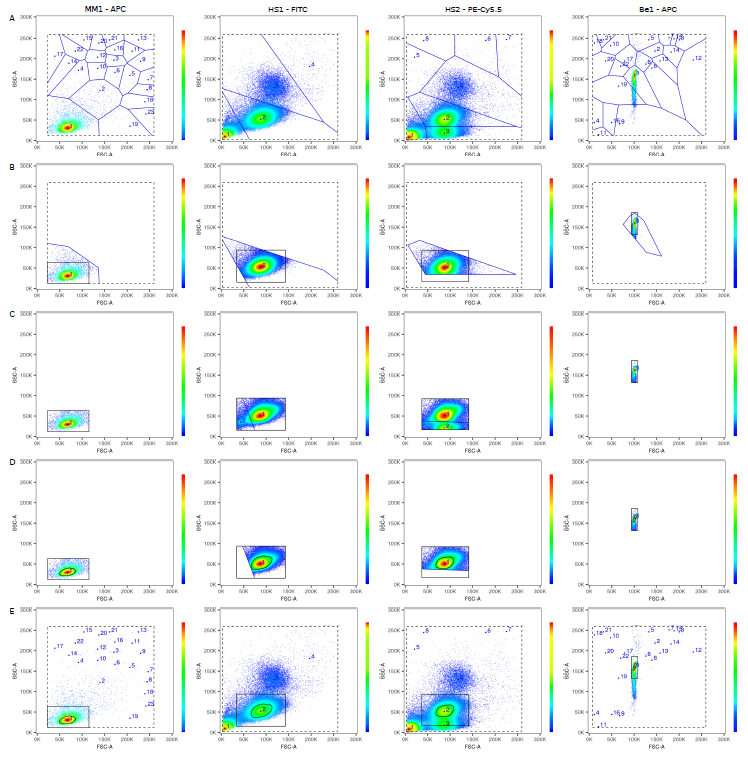
1. Draw an automatic live cell gate
2. Use linear regression to take into account every cell, not just the two data points of average positive and average negative
3. Calculate the error left
4. Tweak the matrix until error is gone
biorxiv.org/content/10.110…
See how easily #AutoSpill removes this autofluorescence specifically from the macrophages:
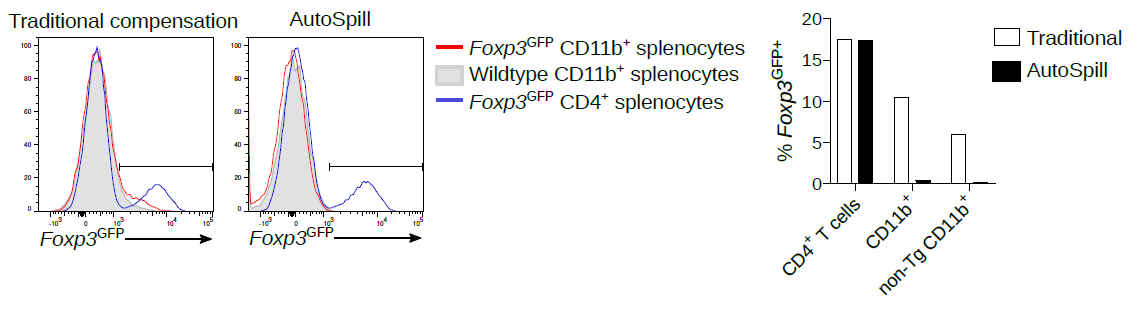
❤️#flowcytometry
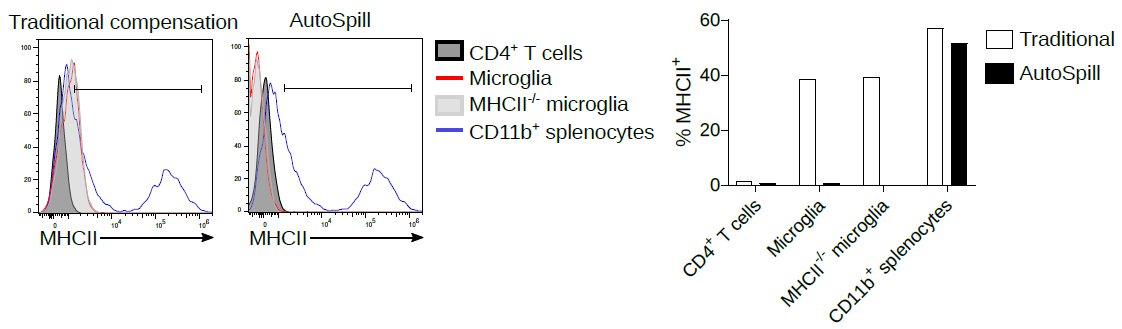
liston.babraham.ac.uk/blog/2020/6/18…
❤️Core Facility staff❤️