
Following up on general thoughts on antigenic drift of #COVID19 from this weekend, I wanted to discuss what we know about the new variant of SARS-CoV-2 thats emerged in the UK. 1/17
https://twitter.com/trvrb/status/1340409968818671616
This variant is referred to as the B.1.1.7 lineage in cov-lineages.org nomenclature and clade 20B/501Y.V1 in @nextstrain nomenclature and can be seen here within circulating viral diversity, where the variant lineage is highlighted in orange (nextstrain.org/ncov/europe?c=…). 2/17 
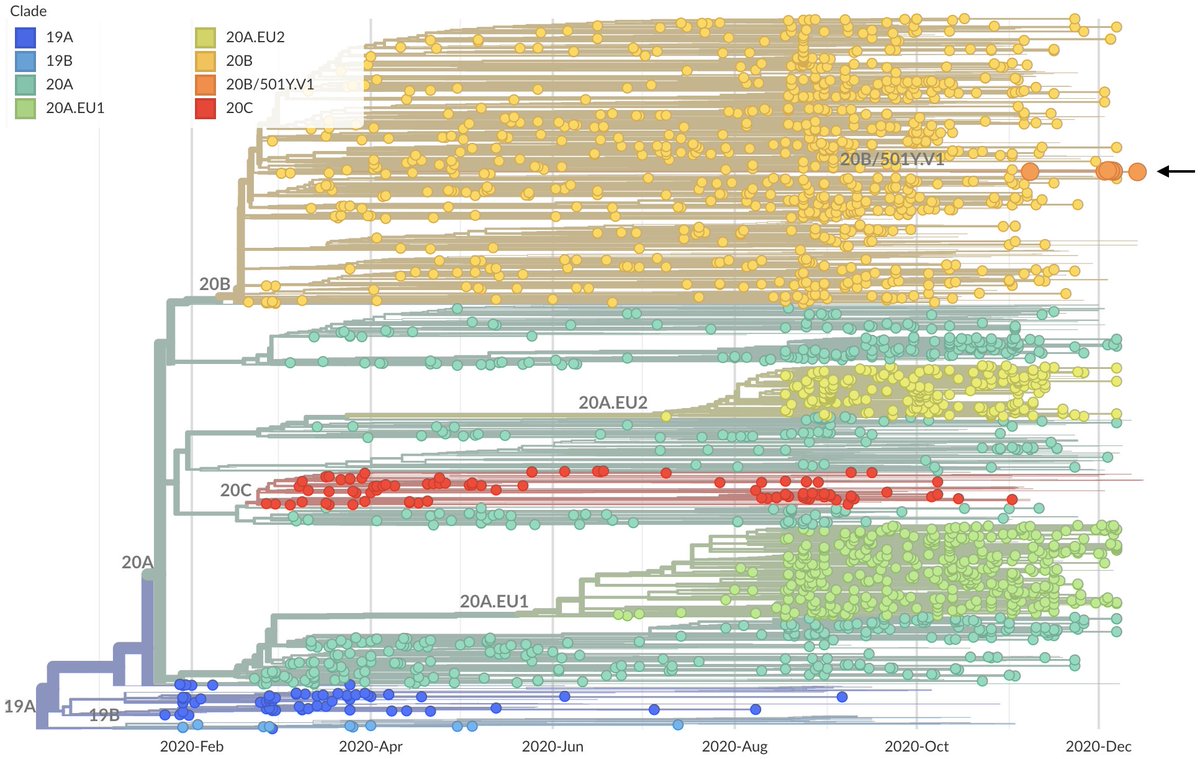
Broadly, I'd characterize the source of concern as arising from the combination of:
1. Multiple mutations that from sequence composition alone are suggestive of biological importance
2. Observed rapid epidemic spread
3/17
1. Multiple mutations that from sequence composition alone are suggestive of biological importance
2. Observed rapid epidemic spread
3/17
For point 1 (mutational composition), the best reference is @arambaut, @pathogenomenick et al (virological.org/t/preliminary-…), but briefly there are 4 different mutations that are particularly striking. 4/17
These are:
1. Spike N501Y which alters a key residue responsible for binding to the human ACE2 receptor
2. Deletion of spike residues 69 and 70 which may aid antibody escape
3. Spike P681H which is adjacent to the furin cleavage site
4. Knockout of accessory gene ORF8
5/17
1. Spike N501Y which alters a key residue responsible for binding to the human ACE2 receptor
2. Deletion of spike residues 69 and 70 which may aid antibody escape
3. Spike P681H which is adjacent to the furin cleavage site
4. Knockout of accessory gene ORF8
5/17
Many of these mutations have been characterized in the lab and we know something about them. However, their combination (along with other changes) has not been characterized. 6/17
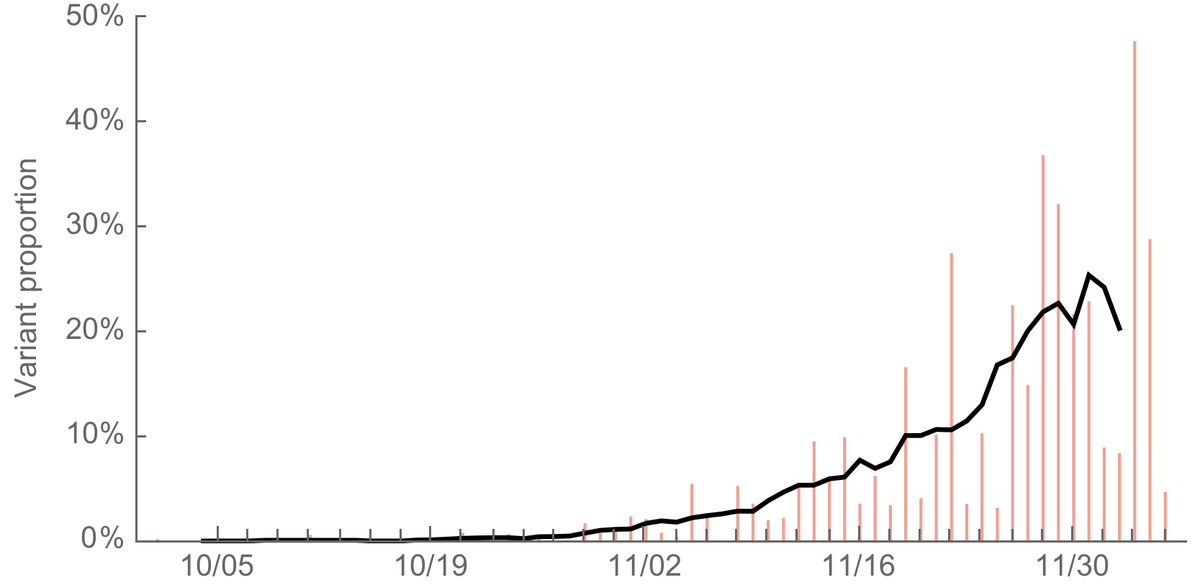
For point 2 (rapid spread), we know the variant emerged recently (~Sep) and has quickly come to comprise a large fraction of cases sequenced by @covidgenomicsuk. Here, I'm plotting daily proportion of sequenced cases that comprise the variant and a 7-day average. 7/17 
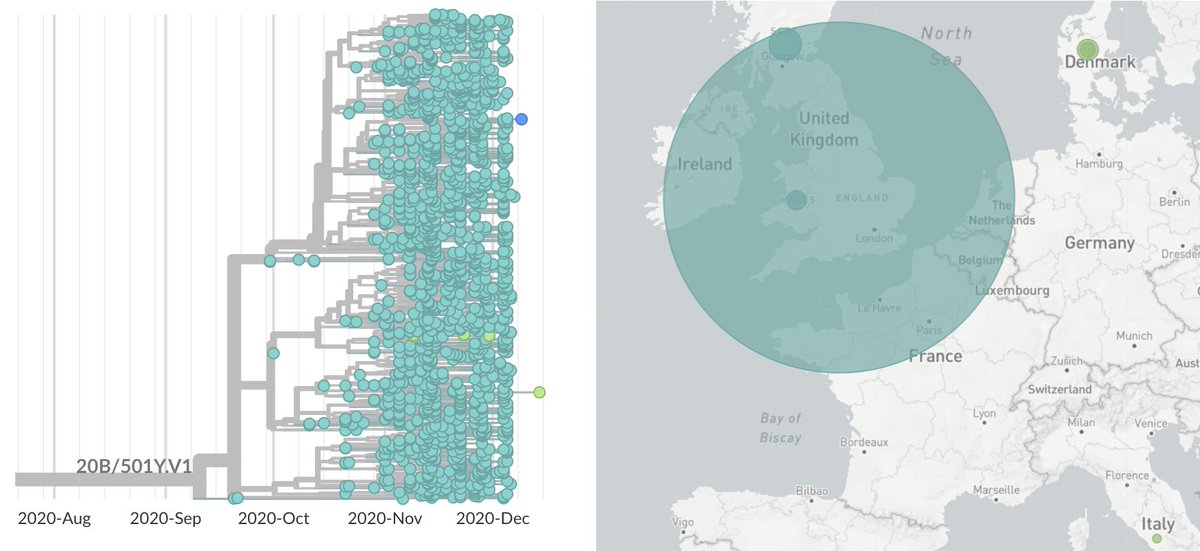
If we look at a detailed phylogenetic tree of this variant, rapid spread is also apparent. We also see that almost all the 501Y.V1 cases are in the UK, with 1 sequenced case from Australia (quarantined), 1 from Italy and 9 cases from Denmark (nextstrain.org/groups/blab/nc…). 8/17 
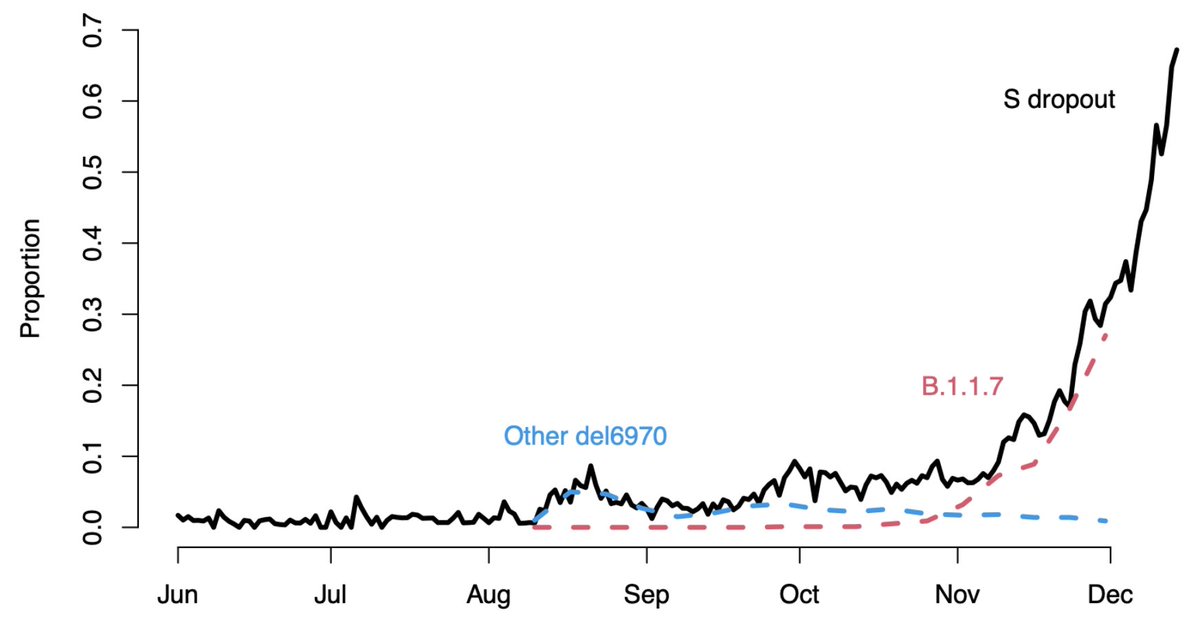
Fortuitously, the spike deletion at residues 69 and 70 broke one of the three PCR probes used in the TaqPath assay and so @PHE_uk has been able to track spread of variant by comparing tests where all three probes light up vs tests where only two light up. 9/17
Here, I'm borrowing a figure from the recent @PHE_uk technical report (assets.publishing.service.gov.uk/government/upl…) showing a dramatic increase in proportion of tests with "S dropout" indicative of a variant case. 10/17 
The dramatic increase in frequency of the variant has been associated with increases in overall epidemic growth as measured by R in regions where it's circulating as described in the PHE technical report and the NERVTAG meeting report (khub.net/documents/1359…). 11/17
In addition to the primary factors 1 (mutational composition) and 2 (rapid spread), we also see higher viral loads in 501Y.V1 cases with Ct values on average 2 units lower. 12/17
Note that "70% more transmissible" is based entirely on the rapid predominance of the variant over existing diversity. However, this evidence is strong enough that I believe a biologically-driven increase in transmissibility is the most likely hypothesis for the data. 13/17
The mechanism for this increase could be higher viral loads and infected individuals being consequently more contagious, but it could also be due to things like increased presymptomatic period or more asymptomatic infections that maintain viral load. We just don't know. 14/17
We don't yet have data on changes in severity. This could be established by comparing frequency of 501Y.V1 in all detected cases vs frequency of 501Y.V1 in hospitalized cases. I'm sure @PHE_uk is working on this now. 15/17
We also don't know if this variant will alter antigenicity. I believe that large reductions in vaccine efficacy are unlikely, but that a modest effect is possible. Experimental work conducted with post-vaccination sera will shed light on this. 16/17
https://twitter.com/trvrb/status/1340410014255652865
Finally, we have no evidence of the 501Y.V1 variant in the US at this point. However, the US has sequenced and shared to @GISAID just 37 genomes from specimens collected after Dec 1, while the UK has sequenced and shared 3774. So, we can't exclude some limited circulation. 17/17
• • •
Missing some Tweet in this thread? You can try to
force a refresh






