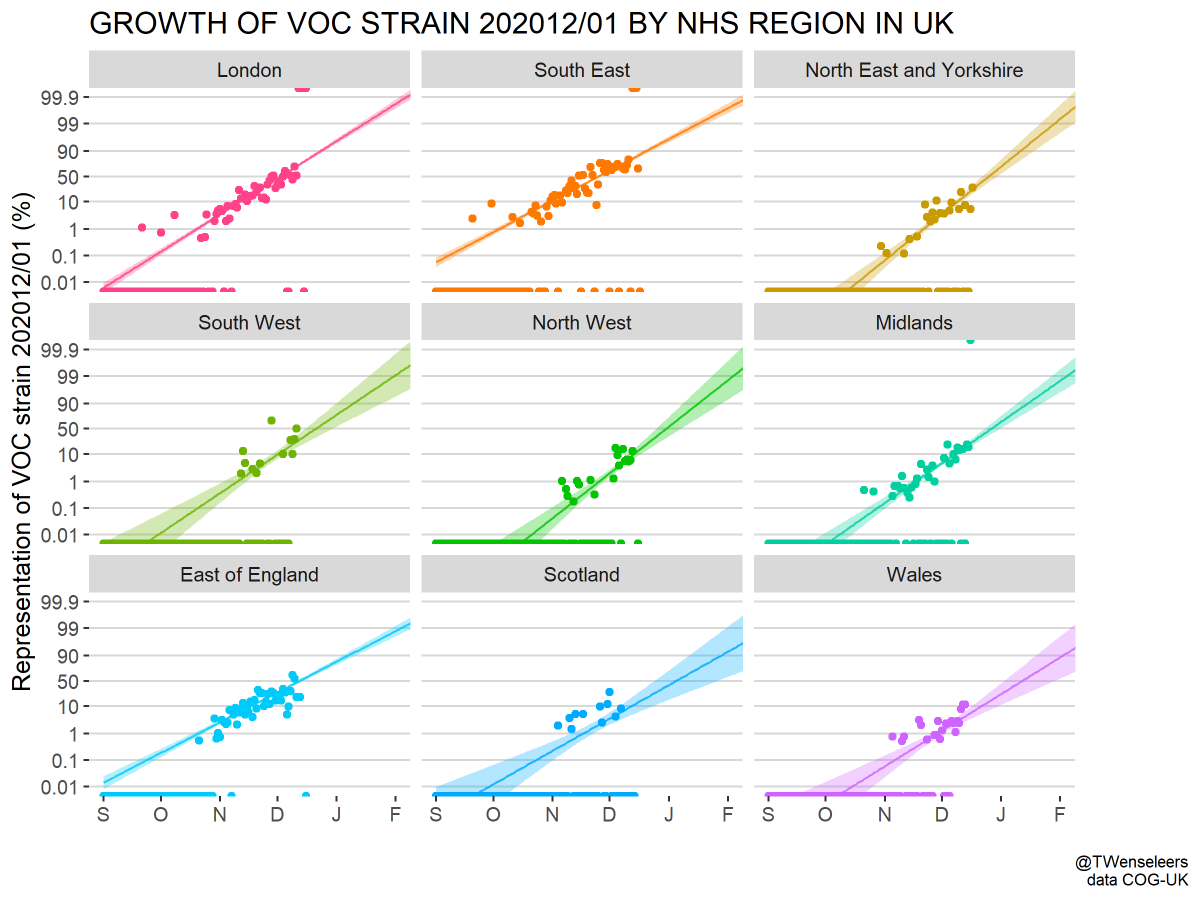
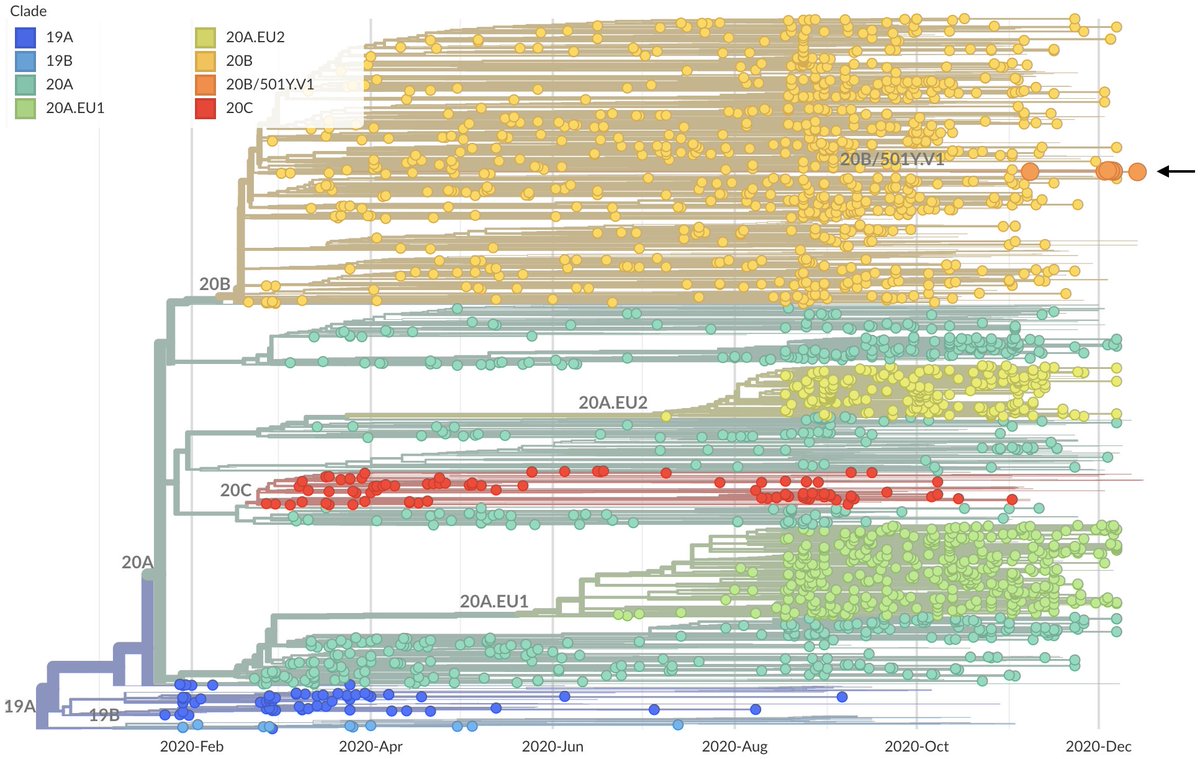
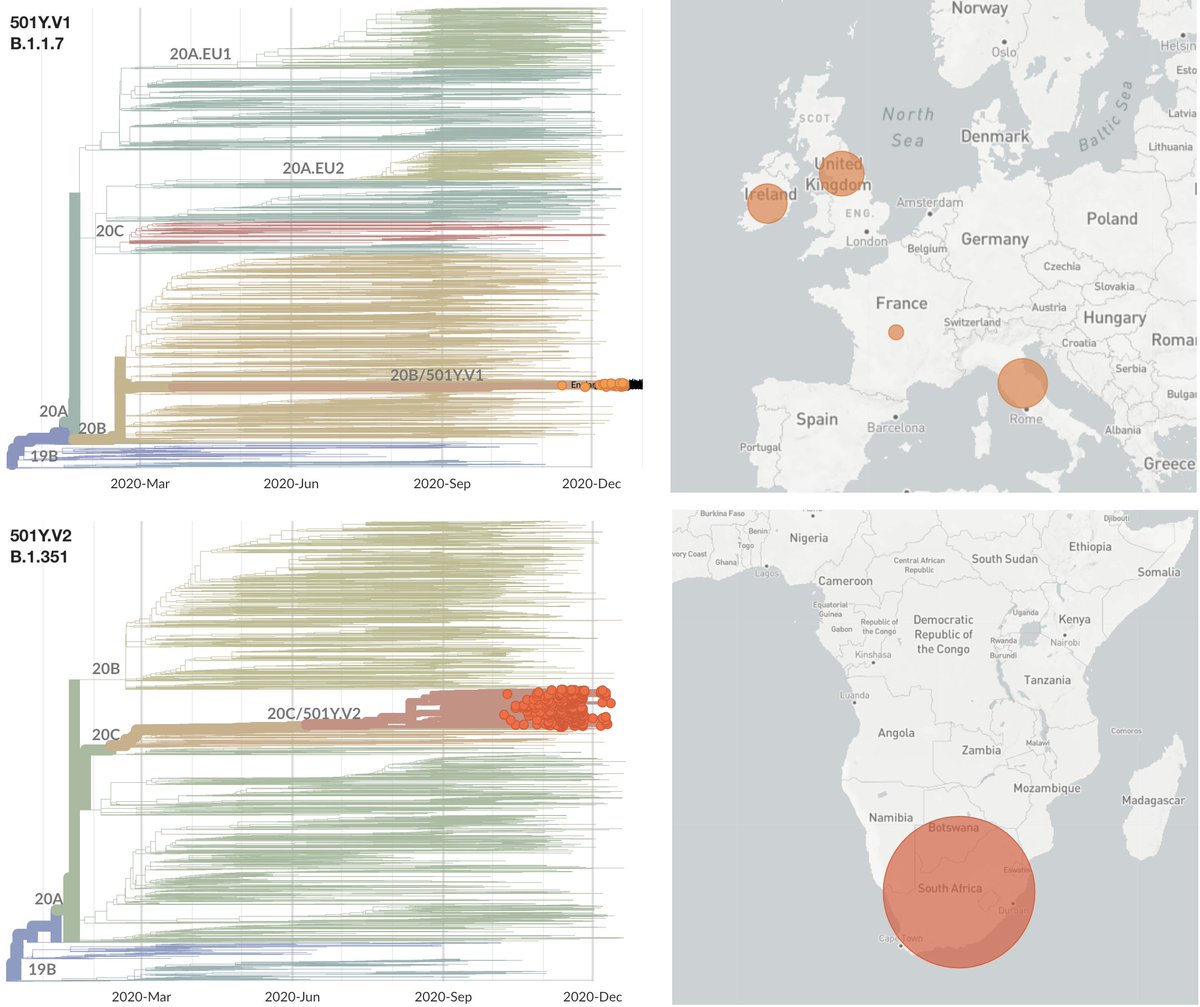
Given that the US has not detected SARS-CoV-2 variant viruses 501Y.V1 or 501Y.V2, what bounds can we place on their current frequency in the US based on sample counts? 1/12
https://twitter.com/trvrb/status/1341446447045087232
So far, the US has not detected any cases that genetically match either the UK variant virus 501Y.V1 (nextstrain.org/ncov/europe?c=…) or the South African variant virus 501Y.V2 (nextstrain.org/ncov/africa?c=…). 2/12 
However, because the US is generally slower at turnaround of specimens into SARS-CoV-2 sequences than the UK, we lack confidence that 501Y.V variants are absent from the US. 3/12
https://twitter.com/trvrb/status/1341806647006543872
Here, I'm using SARS-CoV-2 sequence data available in @GISAID as of last night, and this plot is showing specimen collection date of genomes available from the US. The paucity of Dec relative to Oct and Nov is due to turnaround time from specimen collection to sequencing. 4/12 

We can use a Bayesian approach to estimate the 95% uncertainty interval of observing 0 variant viruses in n sequenced cases (en.wikipedia.org/wiki/Binomial_…). 5/12
Using this method, observing 0 variant viruses in 112 specimens collected in December yields a 95% uncertainty interval of 0% to 2.2% frequency. These December specimens are primarily from California, Michigan, Minnesota and Utah. 6/12 

Importantly, thanks to rapid work by @lapublichealth, 24 of these California samples are from Los Angeles. Observing 0 out of 24 501Y.V samples from LA strongly suggests that the recent epidemic surge in LA is not driven by a more transmissible virus. 7/12
Similarly, observing 0 variant cases in 1776 specimens collected in November yields a 95% uncertainty interval of 0% to 0.14% frequency. These November specimens are from a number of states with the best sampled being Michigan, Minnesota, New York, Virginia and Wisconsin. 8/12 
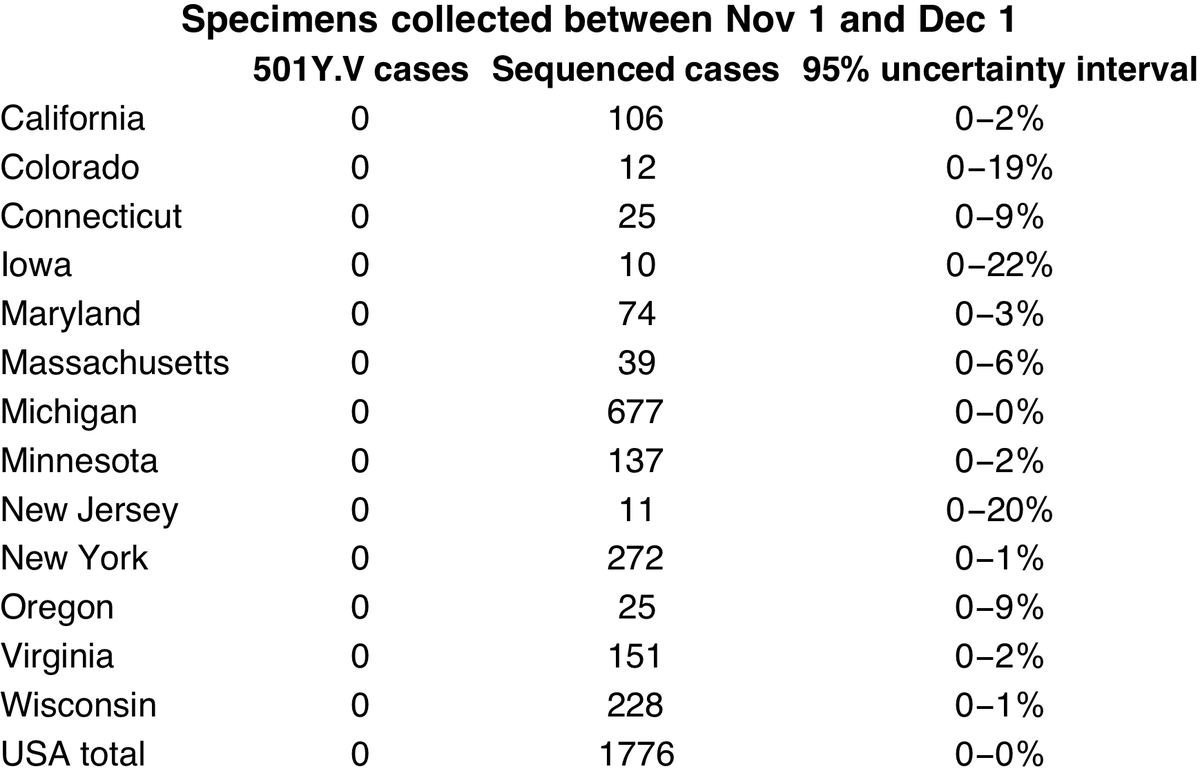
I would conclude that 501Y.V viruses are currently rare in the US if they're circulating at all and are not responsible for recent surges. Geographic sampling in Dec makes conclusions for the US difficult, but data from California (LA in particular) is highly informative. 9/12
This rarity is consistent with @my_helix results showing a slight increase in "S dropout" frequency in Dec of ~0.2% above baseline, again suggesting that 501Y.V1 viruses are currently rare in the US. 10/12
https://twitter.com/alexbolze/status/1341955114043662336
Again, the hypothesized increased transmissibility of 501Y.V is based on epidemiological signals of rapid increase in frequency paired with a signal of higher viral load via Ct value in 501Y.V1. This hypothesis is being intensely investigated. 11/12
https://twitter.com/trvrb/status/1341446478187794432
Huge thanks to @lapublichealth, @K_G_Andersen lab, @michiganhhs, @mnhealth and @UtahDepOfHealth for rapid generation and sharing of the genomic data from specimens collected in December. 12/12
Follow up #1: There is now a sequence confirmed case of 501Y.V1 in Colorado. This was identified through screening of "S dropout" tests. I haven't been able to figure out denominator here. However, we now have 250 sequenced genomes from Dec in GISAID with 0 variant viruses.
• • •
Missing some Tweet in this thread? You can try to
force a refresh