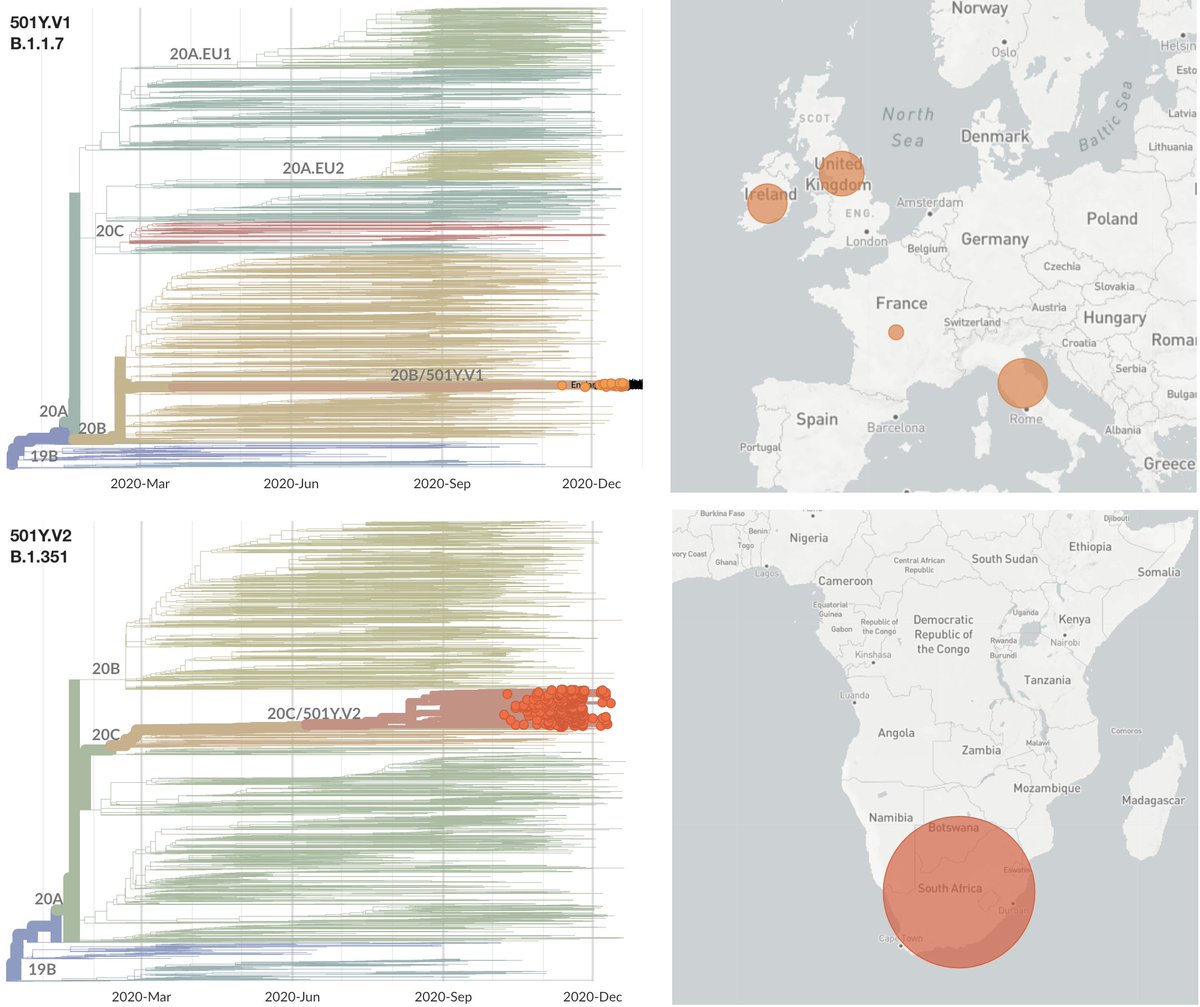
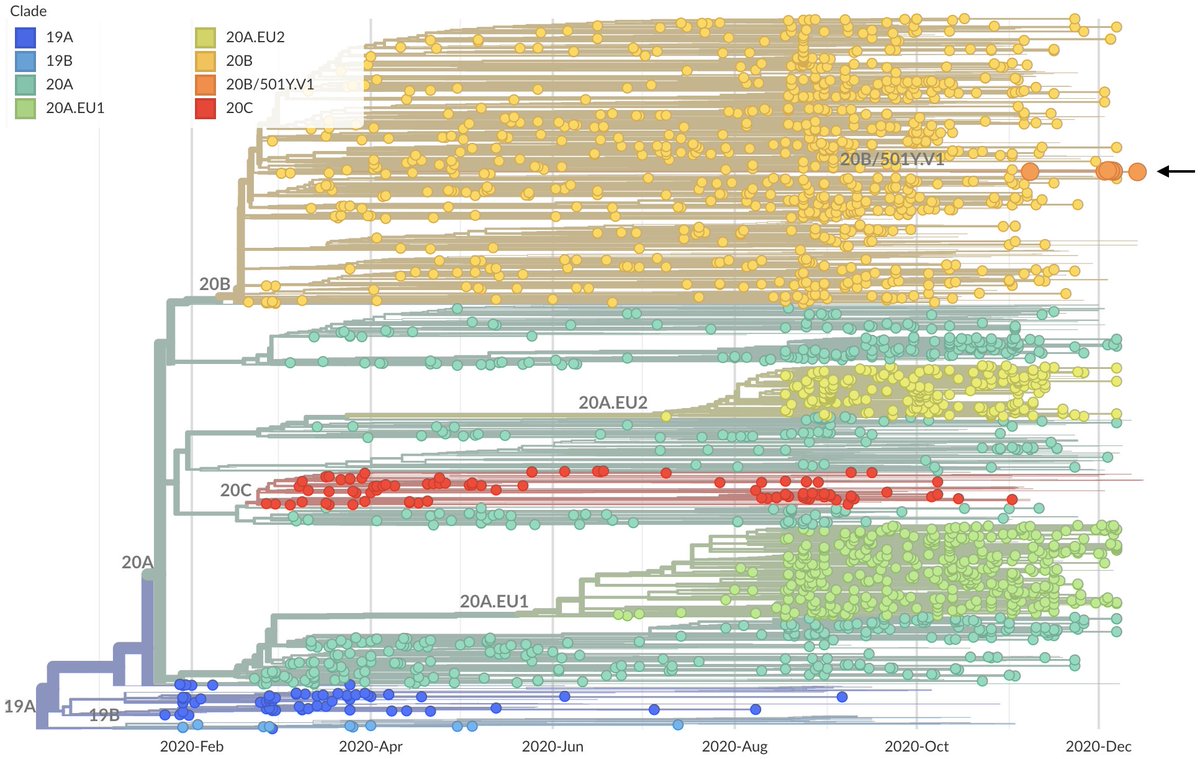
With data that has emerged in the last week, I'm now 80-90% convinced that infections by the UK variant virus (Pangolin lineage B.1.1.7, @nextstrain clade 20B/501Y.V1) result in, on average, more onward infections, ie are more transmissible. 1/10
https://twitter.com/trvrb/status/1341446447045087232
My thinking primarily comes from three data points:
1. rapid increase in frequency of variant over wildtype
2. higher secondary attack rate of variant than wildtype
3. increased viral loads of variant over wildtype as measured by Ct
2/10
1. rapid increase in frequency of variant over wildtype
2. higher secondary attack rate of variant than wildtype
3. increased viral loads of variant over wildtype as measured by Ct
2/10
For point 1 (increase in frequency) we have pretty much the same data as of a week ago, where we see increasing frequency of variant over wildtype across the UK. This can be readily seen in this analysis by @TWenseleers. 3/10

https://twitter.com/TWenseleers/status/1343575693544796160
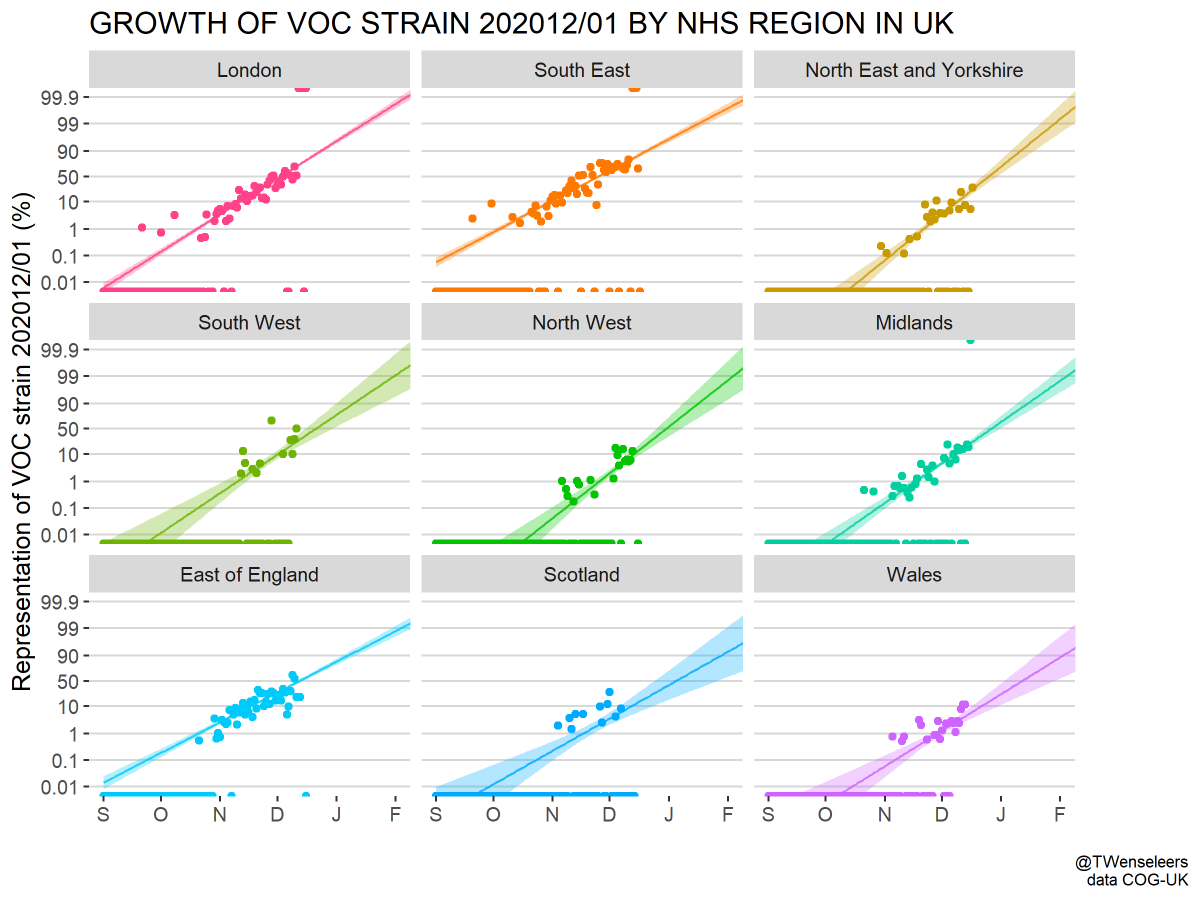
For point 2 (secondary attack rate), we have new data from the second @PHE_uk technical report (gov.uk/government/pub…) where a matched cohort study was conducting comparing 1769 wildtype cases and 1769 matched variant cases. 4/10
By integrating contact tracing data @PHE_uk was able to compare secondary attack rate between wildtype cases and variant cases, where ~10% of contacts of wildtype cases go on to be detected as COVID+ compared to ~15% of contacts of variant cases. 5/10 

Both increase in frequency and secondary attack rate indicate essentially the same quantity, which is the number of secondary infections left by a primary infection, ie R, and tells us that realized transmission rate is likely higher in variant than wildtype infections. 6/10
For point 3 (viral load), we have new data as reported by Michael Kidd, @alanmcn1 et al (medrxiv.org/content/10.110…) that shows that variant cases (as identified by S dropout) have Ct values ~4 units lower than wildtype cases. 7/10 
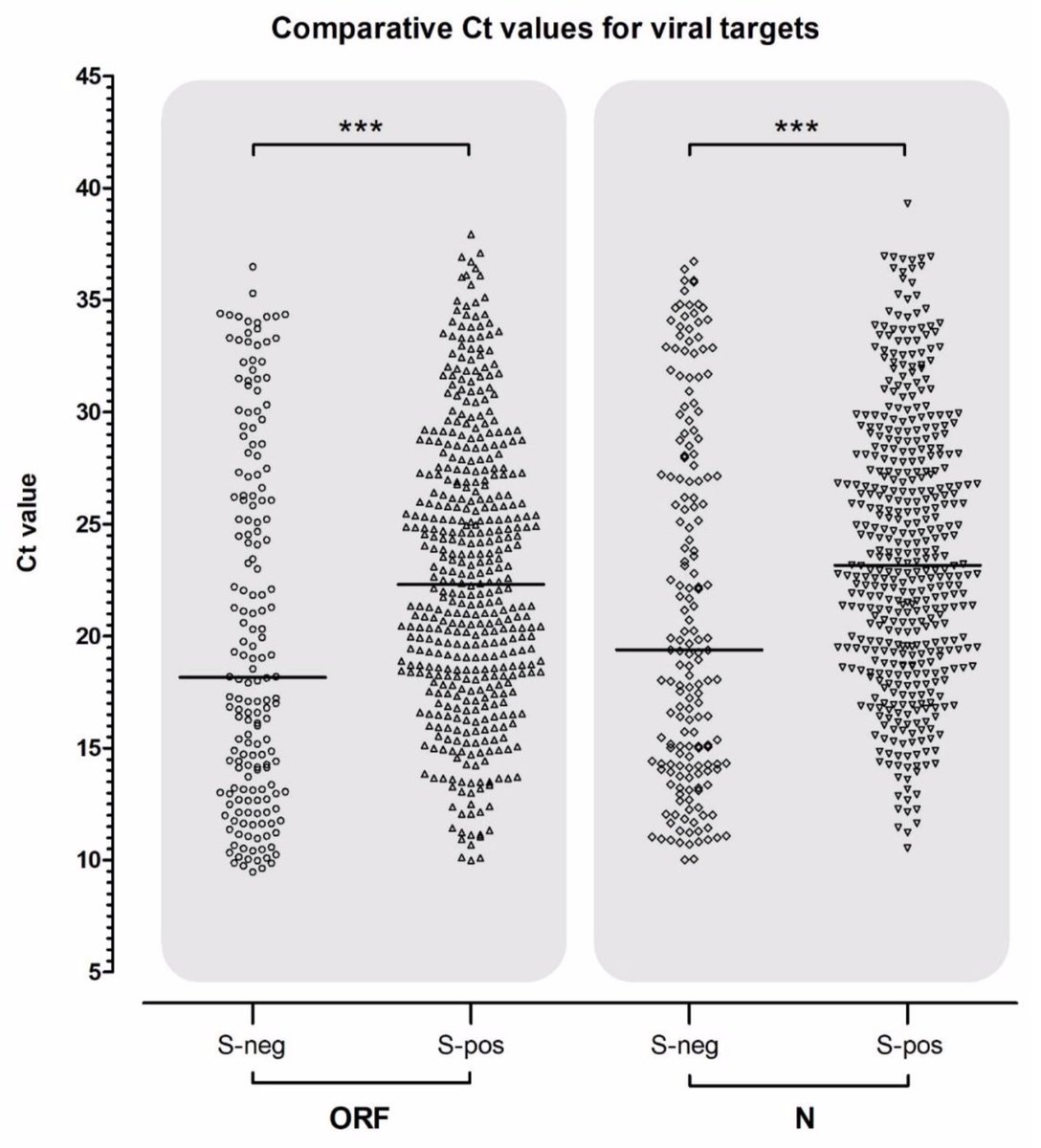
This translates to an estimated 10 to 100-fold increase in average viral load of variant cases. This gives a hypothesized mechanism for increased transmissibility, ie individuals infected by the variant will on average expel more virus and be consequently more infectious. 8/10
The @PHE_uk matched cohort study shows early evidence for a lack of severity difference in terms of hospitalizations and deaths between wildtype cases and variant cases. And additionally shows lack of detectable differences in re-infection rate between variant and wildtype. 9/10
Scientists in the UK are working intensely to address questions of transmissibility, severity and antigenicity of the variant virus and we should learn more in the coming days and weeks, and especially as we see how the UK epidemic continues to play out. 10/10
Follow up #1: The @PHE_uk secondary attack rate analysis was not done on the matched cohort. There should be more stratification here. I'm sorry for the confusion. I'd take the 15% vs 10% secondary attack rate with a grain of salt for the moment.
https://twitter.com/SMHopkins/status/1344006233024585728
• • •
Missing some Tweet in this thread? You can try to
force a refresh