We mapped how all mutations to #SARSCoV2 receptor-binding domain (RBD) affect recognition by convalescent polyclonal human sera (biorxiv.org/content/10.110…).
Among implications: E484K (South African lineage) worrying for immune escape; RBD mutations in UK lineage less so (1/n).
Among implications: E484K (South African lineage) worrying for immune escape; RBD mutations in UK lineage less so (1/n).
We first determined where in #SARSCoV2 mutations most affect viral neutralization. @veeslerlab had reported RBD-binding antibodies responsible for most neut activity of human sera: sciencedirect.com/science/articl…. We validated w sera from @HelenChuMD's HAARVI cohort (below) (2/n) 

Since RBD is main antigenic region (although NTD also important, see below), @AllieGreaney applied method she & @tylernstarr developed for monoclonal antibodies (sciencedirect.com/science/articl…) to map how all mutations to RBD affect binding by *polyclonal* human sera (3/n)
Below are escape maps for sera of 4 people (preprint has more). Tall letters mean mutations greatly reduce serum binding. 3 main sera epitopes in RBD. Most important centers on E484 (dark blue), another is 443-450 loop (light blue), last is distal from ACE2 (orange) (4/n) 

But lots of variation in specificity of anti-RBD immunity among people. For instance, image in previous tweet shows subjects C (& to lesser degree I) focus on E484, subject G focuses on 443-450 loop, and subject K has broad binding not much affected by any single mutation. (5/n)
Key results validate in neutralization assays. Mutations at E484 greatly reduce neutralization (>10 fold) by sera from some individuals (examples below). Unfortunately, the South African lineage has E484K, as do some other isolates from elsewhere. (6/n) 

But as escape maps show, effects of mutations differ across individuals. For subject G (below), serum neutralization not much affected by E484K, but reduced >10-fold by G446V. And some sera not much affected by any RBD mutation (eg, subject K in 4th tweet in thread). (7/n) 

Also, sometimes effects of mutations change over time for same individual as immune response matures. For subject G below, mutations in 443-450 loop cause big reduction in serum antibody binding at day 18 post-symptom onset, but much less effect several months later. (8/n) 

Here are neutralization assays validating maps in prior tweet: G446V greatly reduced neutralization by subject G's sera at day 18, but not day 94 (note total overall neut titers change too).
So heterogeneity both across people and for a single person over time. (9/n)
So heterogeneity both across people and for a single person over time. (9/n)

But can make some generalizations based on examples above + rest of data in pre-print. On average, mutations at E484 have biggest effect on polyclonal sera. Also large effects at sites structurally adjacent to E484 (pre-print has list), and in 443-450 loop for some sera. (10/n)
These results broadly consistent w other studies that selected RBD mutations that greatly reduce neut by polyclonal sera. See papers by @PaulBieniasz @theodora_nyc @vsv512 @e_andreano @McLellan_Lab: biorxiv.org/content/10.110…, biorxiv.org/content/10.110…, elifesciences.org/articles/61312 (11/n)
We do not see strong antigenic effects of N501Y RBD mutation in UK viral lineage, consistent with these data from @TheMenacheryLab (
https://twitter.com/themenacherylab/status/1341460343168577538). Note that N501Y could affect transmissibility, and UK lineage also has NTD mutations (more on that below). (12/n)
Here's plot of how mutating RBD sites affects average serum binding (y-axis) vs frequency of mutations (x-axis). E484K in S African lineage most worrying. But others affect some serum to various degrees & no such thing as "average" human when it comes to serum specificity (13/n) 
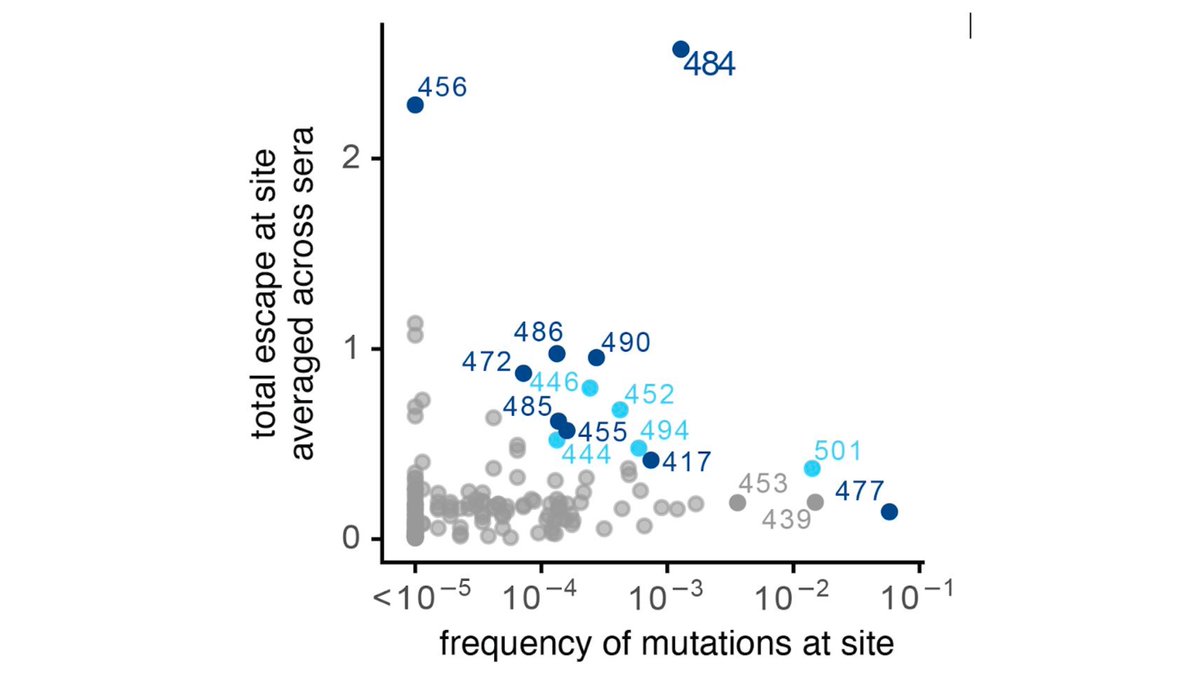
Importantly, we only looked at RBD muts, since majority of neut activity of most sera from RBD antibodies (2nd tweet of thread). But NTD muts also important; see @10queues @mccarthy_kr @GuptaR_lab @e_andreano @McLellan_Lab: biorxiv.org/content/10.110…, medrxiv.org/content/10.110… (14/n)
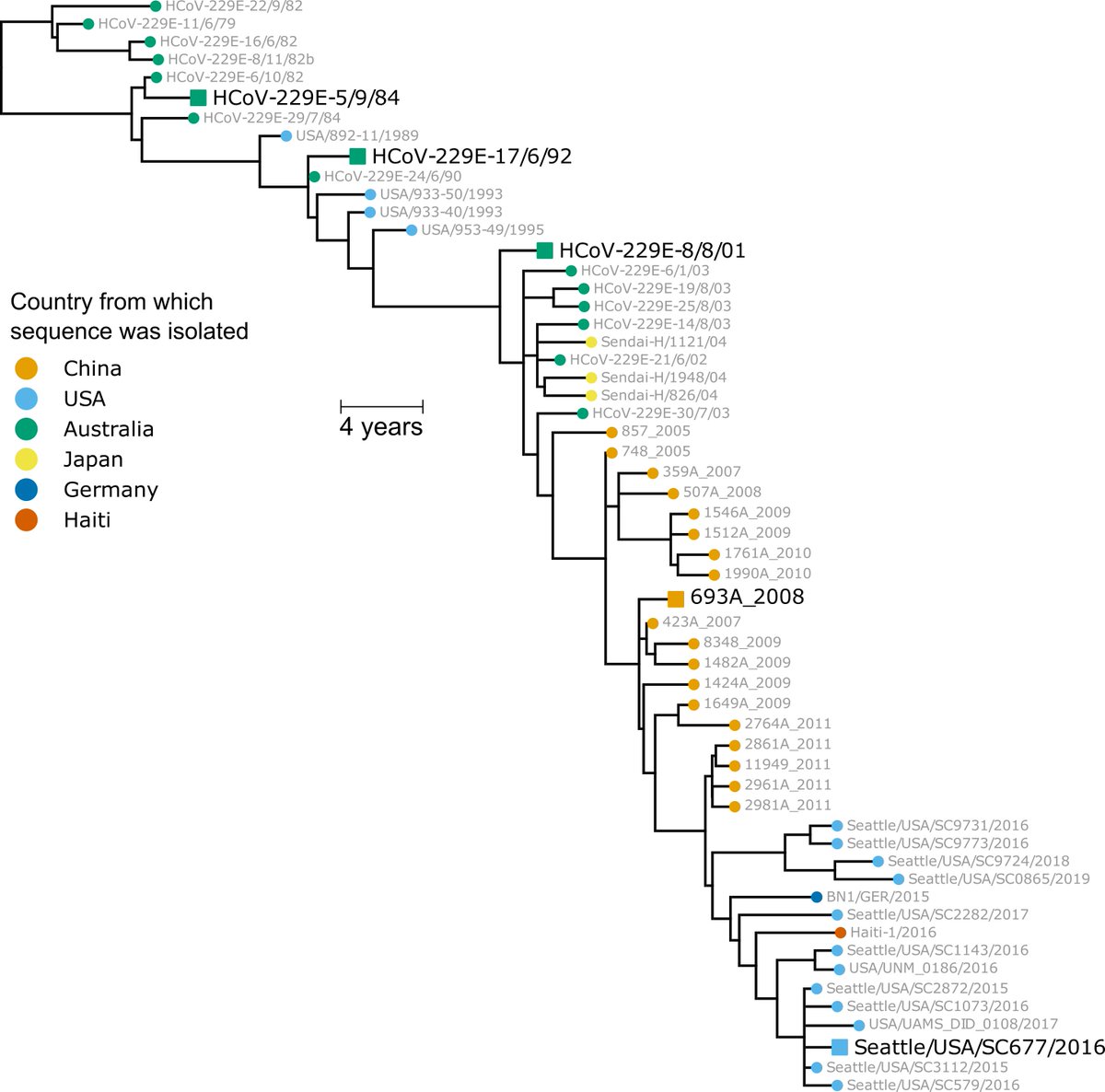
This relative role of RBD & NTD mutations consistent w historical evolution of common-cold CoV-229E, where mutations concentrated in receptor-binding loops of RBD, but also in parts of NTD. Here is plot of mutational variability in CoV-229E spike:
https://twitter.com/jbloom_lab/status/1339939746068353025(15/n)
What do results mean for possible #SARSCoV2 immune escape? Certainly mutations like E484K are concerning. But they *reduce* neut activity, they don't ablate it. Again, look at CoV-229E: takes years of evolution to escape serum neut of most people
https://twitter.com/jbloom_lab/status/1339939732529111040(16/n)
Heterogeneity of mutation effects across sera could be blessing, as mutations that reduce neutralization by some people won't affect others. Look below at how differently evolution of CoV-229E erodes neutralization by different people (Fig 2A of biorxiv.org/content/10.110…) (17/n) 

Heterogeneity also shown by @e_andreano @RommieAmaro @McLellan_Lab, who found multiple mutations that greatly (300-fold) reduce neut of #SARSCoV2 by one human serum but often only modestly (a few fold) affect other sera: biorxiv.org/content/10.110… (18/n)
Scientifically, we see two priorities going forward: (a) Perform similar studies of vaccine sera. Is vaccine immunity affected by mutations similarly to natural immunity? (b) Monitor for antigenic mutations so vaccines can eventually be updated if needed. (19/n)
But biggest priority is vaccinate! Despite above, I'm confident current vaccines will be useful for quite a while. Reasons: (a) even worst mutations (ie, E484) only erode neut activity of some sera, don't eliminate it for any, (b) current vaccines elicit strong immunity... (20/n)
... (c) evidence in animals (& from humans after 1st vaccine dose) that modest immunity can blunt disease, (d) natural immunity to seasonal CoV provides some homologous protection for 3+ years even though they evolve too (see wellcomeopenresearch.org/articles/5-52) (21/n)
Finally, most credit goes to @AllieGreaney, the outstanding grad student who led our study. She worked with Andrea Loes, @khdcrawford, @tylernstarr, & Keara Malone. @HelenChuMD was our invaluable clinical collaborator, and thanks to the volunteers in her HAARVI cohort (22/n)
I'd also like to call out the important work by @houzhou @Tuliodna and cowoerkers in characterizing lineage 501Y.V2 initially identified in South Africa, which carries RBD mutations including E484K, which our study suggests has an antigenic effect: medrxiv.org/content/10.110… (23/n)
Our functional work is potentially useful because the high-quality viral surveillance and rapid reporting of data exemplified by their study.
I'd also like to correct terminology earlier in this Tweet chain. It is lineage 501Y.V2, initially identified in South Africa. (24/n)
I'd also like to correct terminology earlier in this Tweet chain. It is lineage 501Y.V2, initially identified in South Africa. (24/n)
It should be referred by full lineage, not country as in my earlier Tweet. In addition, E484K has been independently observed in viruses from other locations, so any antigenic impacts of this mutation is *not* a concern confined to any country or viral lineage. (25/n)
One more Tweet to share data by @firefoxx66 inspired by useful comments from @Tuliodna. Please read this:
Key implication: the E484K mutation was most prominently reported by @Tuliodna in the 501Y.V2 lineage first identified in South Africa. (26/n)
https://twitter.com/firefoxx66/status/1346546912644759553?s=20
Key implication: the E484K mutation was most prominently reported by @Tuliodna in the 501Y.V2 lineage first identified in South Africa. (26/n)
But importantly, muts at E484 arisen multiple times in several countries over last 9 months. Our functional work (& others cited above) show E484 muts antigenically important. And surveillance by @Tuliodna @rjlessells @atrvlncc & others show this mutation is circulating (27/n)
But because this mutation has arisen multiple times in multiple locations, it is not a concern confined to any particular country. Instead, globally important to continue to monitor evolution, characterize mutations, & assess any eventual implications for vaccine updates. (28/n)
• • •
Missing some Tweet in this thread? You can try to
force a refresh