In this short thread, I am going to plot some experimental data in a way that provides perspective on concerns that #SARSCoV2 mutation E484K will completely abolish immunity. (Thanks @profshanecrotty @apoorva_nyc for inspiring this post.) (1/n)
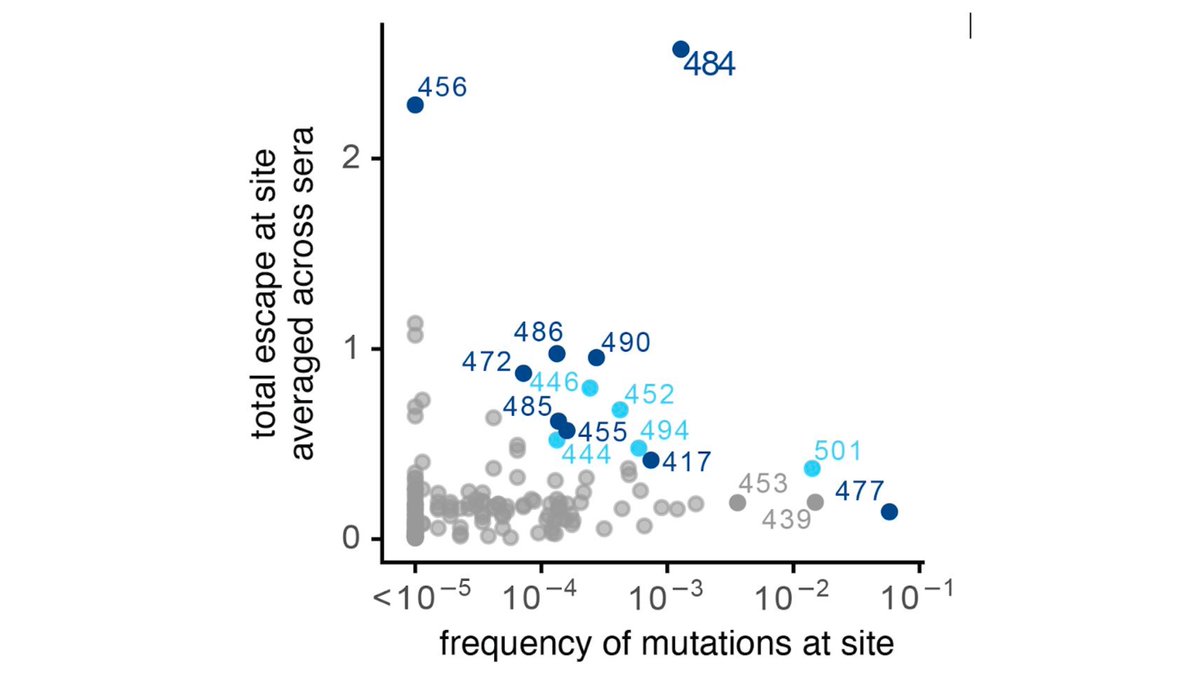
Last week, we posted a study describing how some #SARSCoV2 mutations, especially at site E484, reduce binding & neutralization (
https://twitter.com/jbloom_lab/status/1346442000472580098). This study (& similar ones by other) have drawn a lot of interest since E484K is in B.1.351 viral lineage. (2/n)
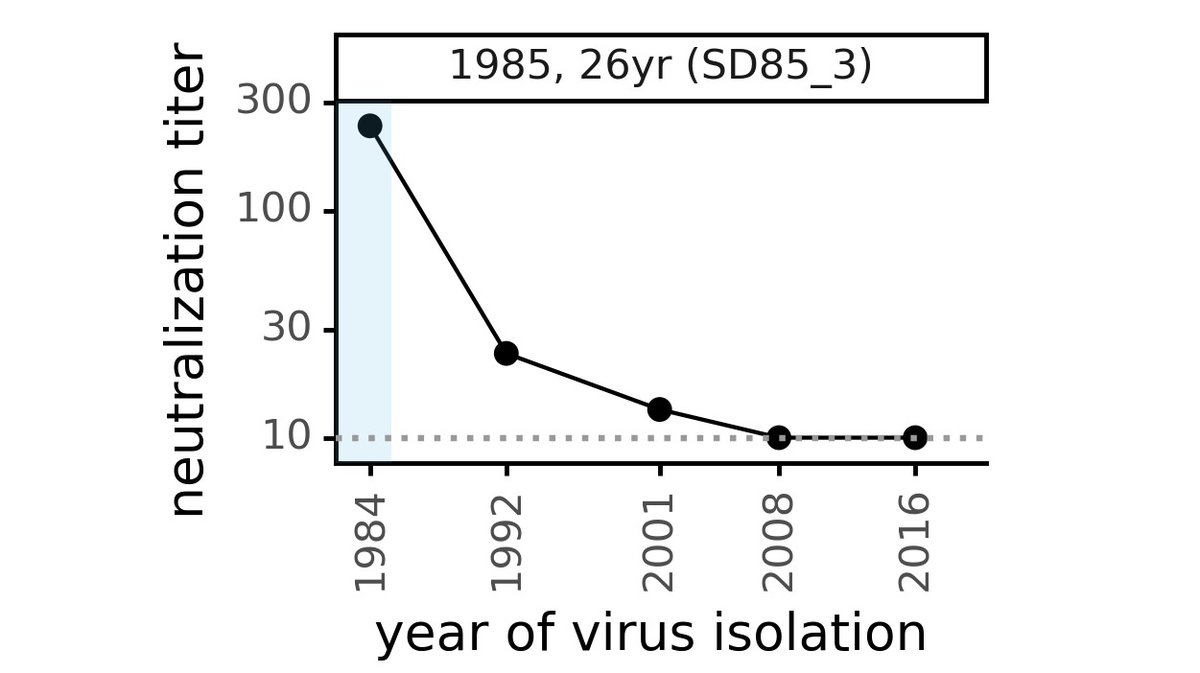
However, E484 mutations *reduced* neutralization, they did not ablate it. The plot below shows how E484 reduces neutralization titers for 16 sera. The dashed orange line shows titers against unmutated virus (measured by Pfizer) after 1 dose of BNTB162 vaccine. (3/n) 

Plot in previous tweet shows E484 mutations often reduce neutralization (lines slope down). Sometimes effect is big (eg, subject C, day 32). But the sera we tested still neutralized E484 viruses, at titers well above those to unmutated virus after 1 dose of Pfizer vaccine. (4/n)
If you recall 2 weeks ago, Twitter was buzzing with people saying "1 vaccine dose is enough, just give that." It puts E484 in perspective to note these mutant viruses still neutralized at a titer greater than what many people were advocating as "good enough" quite recently. (5/n)
Should we worry about E484K & other mutations? Yes! That's why so many of us are working hard to study them. But we need to keep perspective. Reduced neutralization does not mean no immunity, & it will take careful study to determine implications for protection in humans. (6/n)
Finally, results from my lab described above are consistent with work of many other groups. Specifically... (7/n)
@e_andreano @RommieAmaro @McLellan_Lab selected virus w E484K (+ more muts) that was 300-fold more resistant to one human serum. But was still neutralized by that sera & most others they tested, often w titers dropping by just 2-5 fold (Table S1 of biorxiv.org/content/10.110…). (8/n)
@theodora_nyc @PaulBieniasz found E484K reduced neutralization by several human sera, but in all cases sera still neutralized mutant virus, sometimes quite potently (Figure 6C of elifesciences.org/articles/61312). (9/n)
@vsv512 found mutations at E484 often reduced serum neutralization, but every sera they show in Figure S5 of biorxiv.org/content/10.110… still neutralizes the mutants to at least some degree. (10/n)
So we need to monitor these mutations, and be prepared to update vaccines eventually if needed. But we also need to remember that a reduction in neutralization titer, while worrying, is not the same as complete elimination of all immunity. (11/n)
• • •
Missing some Tweet in this thread? You can try to
force a refresh