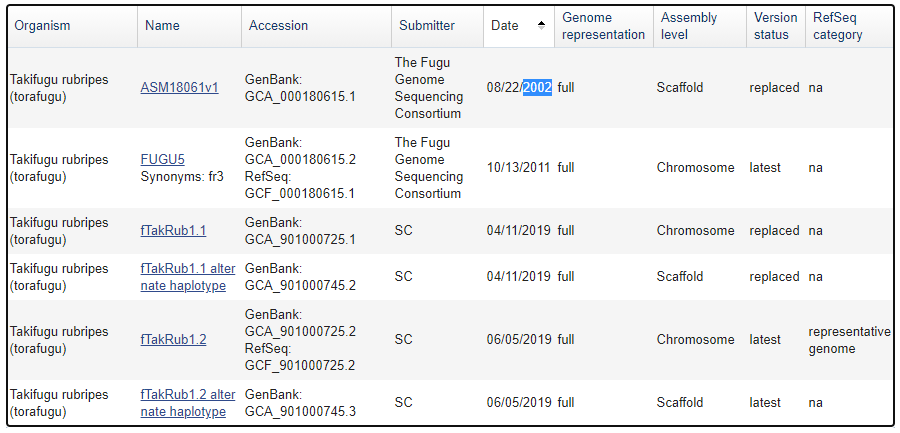
Some stats and facts about the #Takifugu #rubripes assembly by @genomeark: this is the third iteration of the assembly. The first was completed in 2002. There was another iteration done in 2011. Why was the pufferfish sequenced so early? A lot has got to do with Sydney Brenner... 
https://twitter.com/genomeark/status/1349280208277434368
Indeed, as we can see in this archived version of @ensembl, the Fugu genome was the 5th vertebrate genome to be completed, after human/mouse/rat and zebrafish. Even though zebrafish is widely used as a model organism, Sydney Brenner argued that Fugu was worth sequencing ... 

... The reason is that Sydney Brenner was passionate (obsessed?) with gene duplications and functional diversification. I.e. a gene duplicates in two copies, and over time, each copy can specialize in doing something slightly different. ...
... He studied genes related to vision and eye formation. Then why Fugu? He wanted another fish genome, close to zebrafish, but to ease proposition of the millions of $ that would take to sequence (back in 2002) he chose Takifugu rubripes, because something happened to it. ...
Similarly to the chicken genome, the Fugu genome is special in that at some point several million years ago, it "got rid of a lot of junk DNA", shedding out lots of repetitive regions in the genome, not unlike someone going on a diet after Xmas holidays. ...
... So the total genome size of Fugu is much smaller than other similar fishes in the evolutionary tree, and it's kept most of the genes and functional regions, but this smaller leaner genome was a lot cheaper to sequence back in 2002. ...
... So Sydney Brenner got what he wanted: a second fish genome to compare to zebrafish for him to study the fate of gene duplications, but without having to spend as much money as most fish (vertebrate) genomes.
@d2unroll unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh