#JPM2021 @GenapSys My Highlights: I think it's fair to say I am more excited than most about this #NGS company, as I see them as an example of how to enter the market while keeping a small profile ($249M raised so far). 
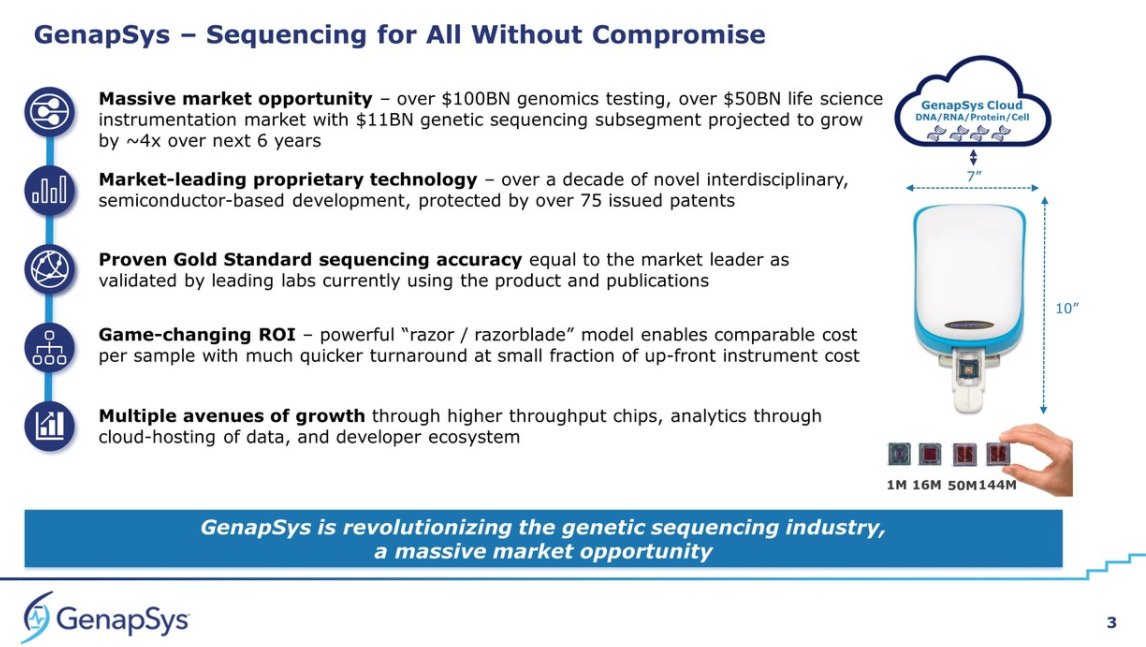
They now are aiming at 2021 to ship two new chips: 50MM read chip and 144MM sensor chip (not sure what the difference is between read/sensor). 

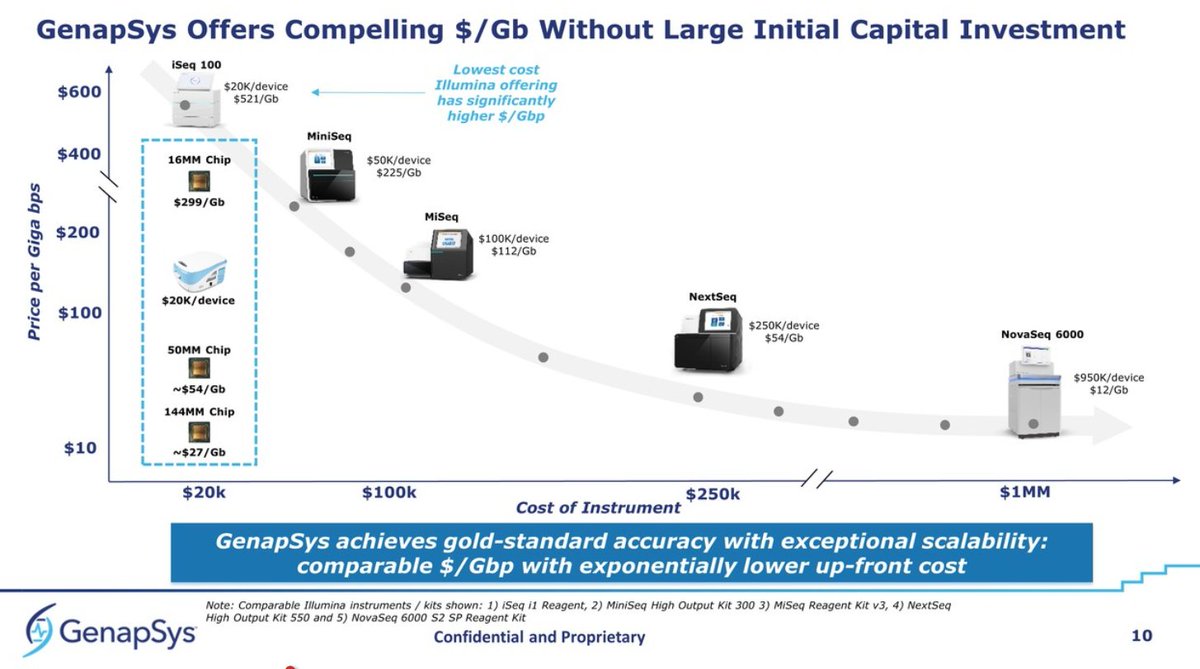
They show a slide of price per Gb with #Illumina products as a reference, and their products now lined up, I think, for the first time with price per Gb info. Lowest will be the 144MM chip at ~$27/Gb. 
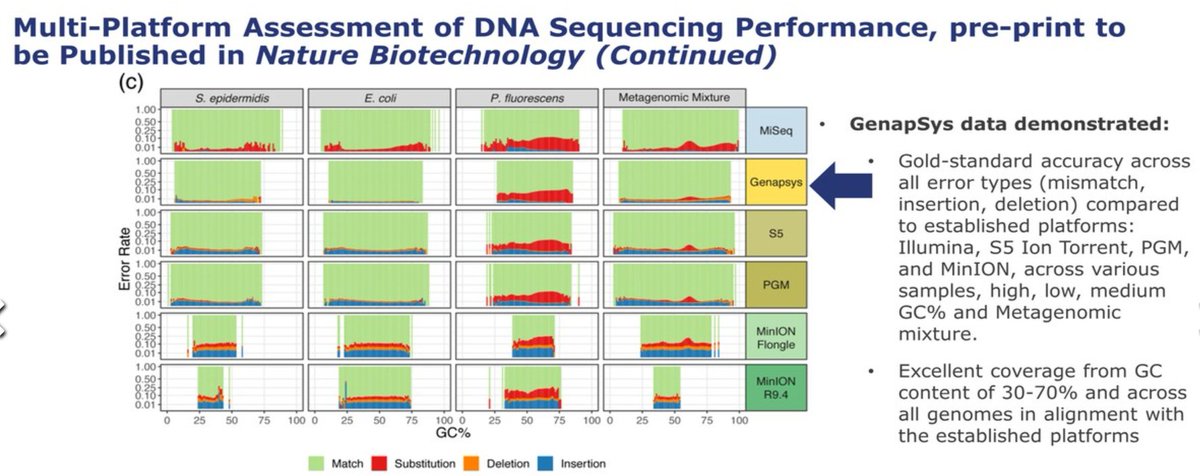
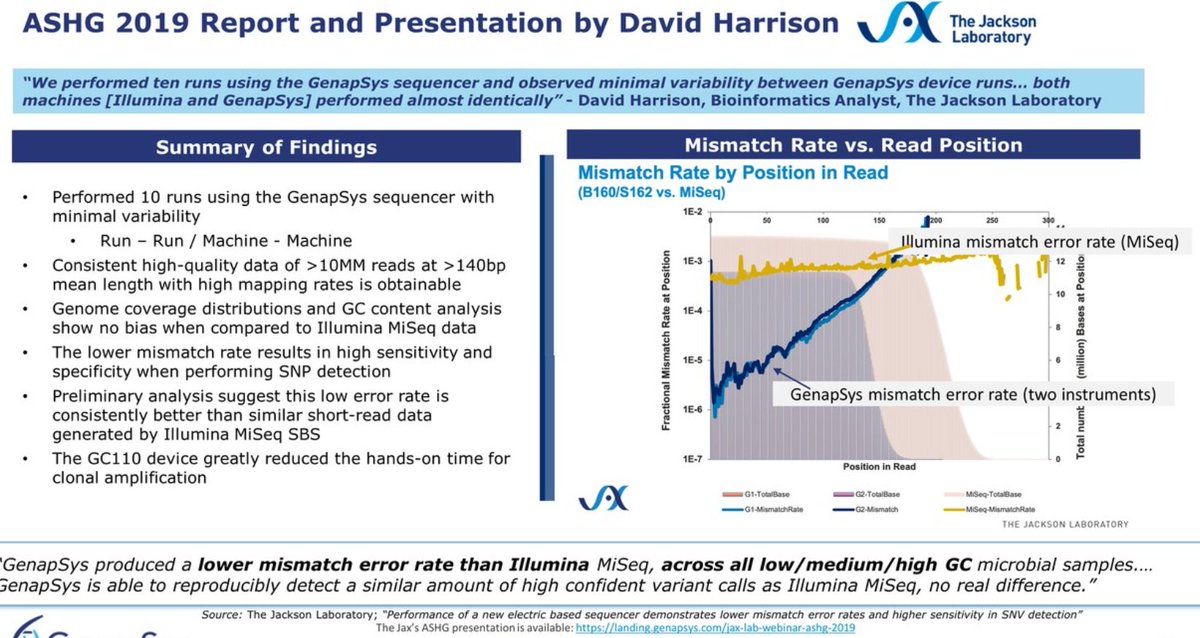
As their technology is closer to the #IonTorrent than to the cycle-by-cycle SBS-based sequencing methods, they should have microindel errors in their profile. These look much better, almost null, than the S5 data they show in comparison. 
The next slide, also about error rates, either for each base as the run progresses or sliced in GC% windows, has a lot of info, but difficult to read. I think the summary is that the @Genapsys profile shows a bit of microindel error but otherwise comparable to 100bp ILMN data. 
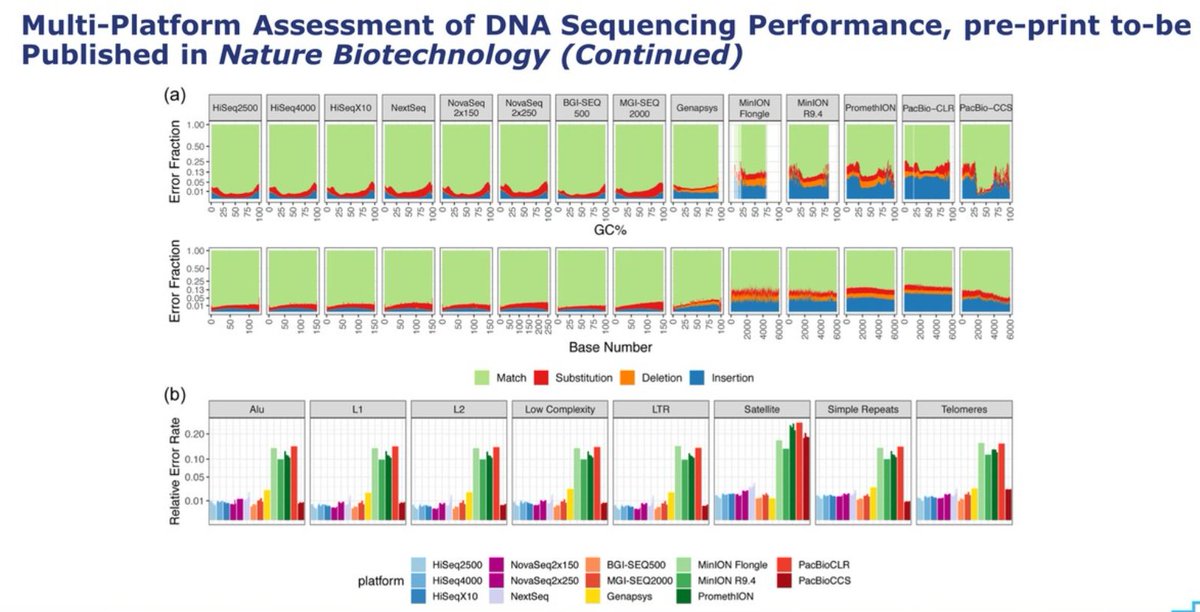
The error profile starts low, very low actually, and then it hikes up after cycle 50-100+. In the slide below, they've been naughty and put a textbox on top of their error blue line, trying to hide it: don't think we won't notice these tricks! 😉 
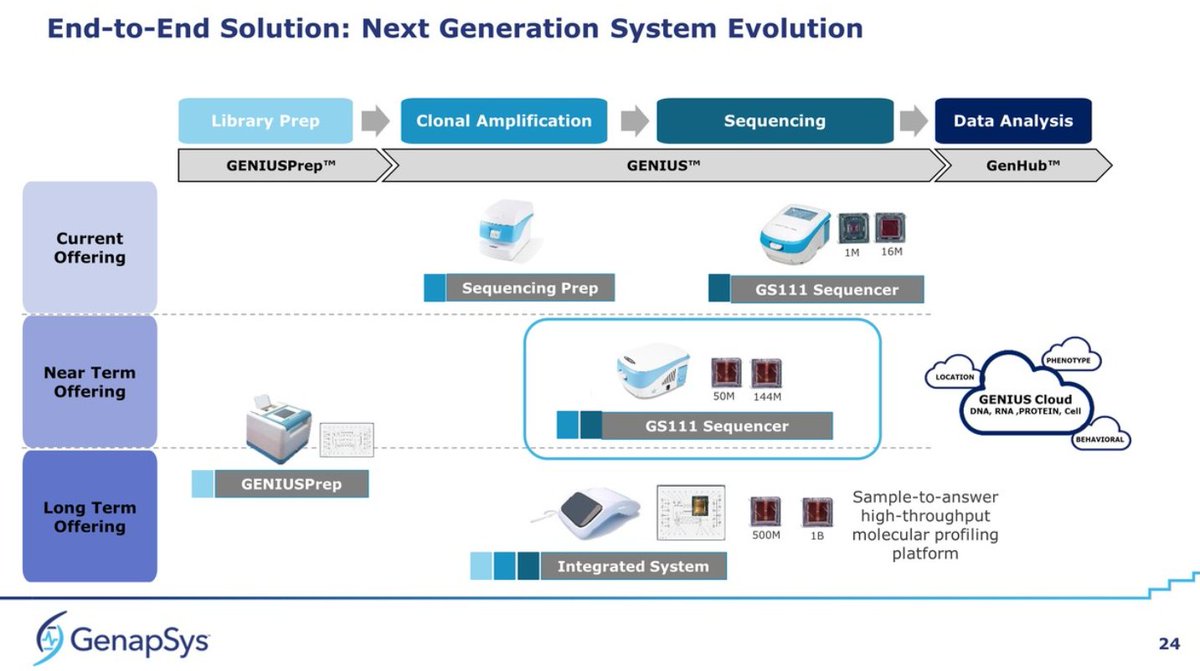
They say they've placed or booked more than 110 instruments so far (does that count early access?). All this is based on a 10K GS111 Sequencer + 10K Sequencing Prep instrument. They show long term plans for a GENIUSPrep (high throughput sample prep), but also ...
Details of the 50M Chip. They seem to have gone from 100bp to 150bp standard now, but raw accuracy still placed at 99.9% at position 125bp. This may be the actual 144M chip but with standard Poisson loading, rendering a bit over 50M reads, or 8Gb per run, which is impressive... 
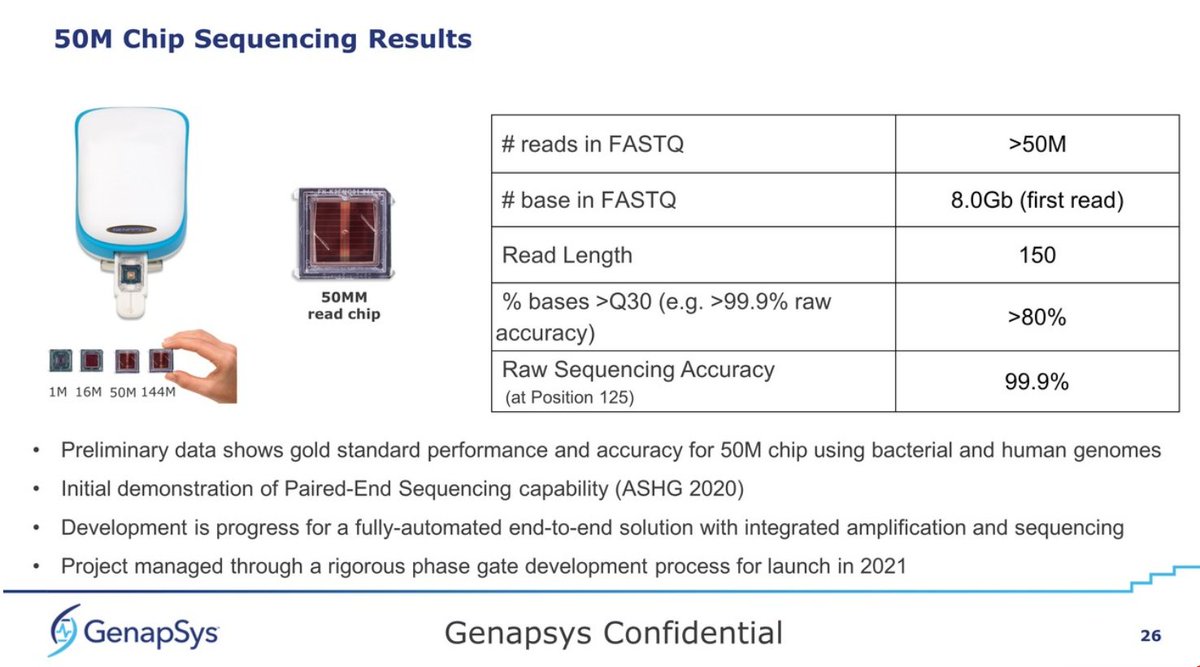
... with a $20K capital investment, comparing the 50M @Genapsys at 8Gb ($54/Gb) and the #Illumina #iSeq100 at 1.2Gb ($542/Gb), that's roughly 8x better yield for Genapsys, and 10x better price per Gb. It makes the #iSeq100 look rather underwhelming in comparison.
The slide about 2021 plans changes the naming from 144M chip to "G4-50M and G4-100M", so this could mean that the nomenclature changes to reads per run rather than the size of the chip. Maybe both based on the 144M chip, but with better loading giving the 50M->100M jump in reads?
No reference to paired-end reads, only indications that 150bp may not be the end of it, and they may be able to go longer than that. Good news about the G4-100M, as it seems they have scope of improvement in loading. 

@d2unroll unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh