Our study mapping #SARSCoV2 mutations that escape key therapeutic monoclonal antibodies is out in @ScienceMagazine. The study also shows that some of these escape mutations arise in a persistently infected patient treated with REGN-CoV-2: science.sciencemag.org/content/early/… (1/n)
I previously summarized the pre-print (
https://twitter.com/jbloom_lab/status/1333897221561978880), so in this thread I'll just update on new insights since we posted the pre-print in late November. (2/n)
In the study, we mapped all mutations to #SARSCoV2 RBD that escape binding by recombinant forms of antibodies in REGN-CoV2 cocktail (Regeneron) and LY-CoV016 antibody (Eli Lilly). These maps are useful because some of these mutations are appearing in new viral lineages (3/n).
Before summarizing key mutations, I'll add that at end of last week @tylernstarr just got data for recombinant form of LY-CoV555 (lead Eli Lilly) antibody. Not sure when we will have time to write paper on that, so I'm posting data below in same form as Figure 1A of paper. (4/n) 
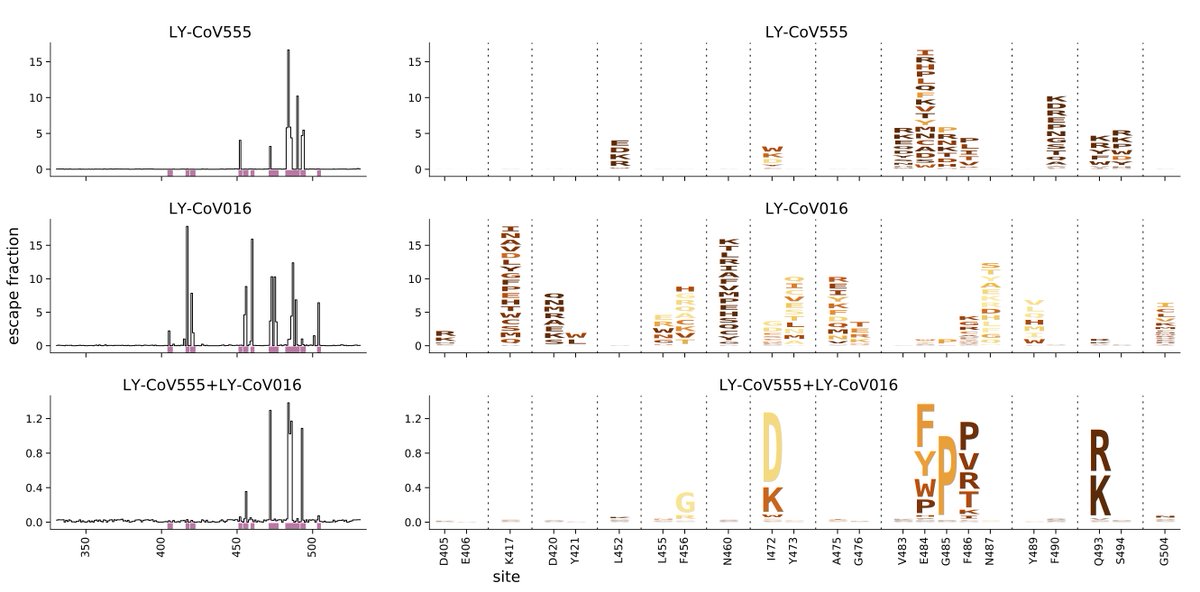
Here are impacts of mutations at some key sites on REGN-COV2 antibodies (REGN10933 and REGN01987), LY-CoV555, and LY-CoV016:
E484: mutations reduce binding by LY-CoV555 and to lesser extent REGN10933, little effect on REGN01987 or LY-CoV016. (5/n)
E484: mutations reduce binding by LY-CoV555 and to lesser extent REGN10933, little effect on REGN01987 or LY-CoV016. (5/n)
N501: mutations have little effect on any of the antibodies.
K417: mutations reduce binding by LY-CoV016 and REGN10933, little effect on REGN1097 or LY-CoV555
L452: mutations reduce binding by LY-CoV555, little effect on REGN10933, REGN01987, LY-CoV016. (6/n)
K417: mutations reduce binding by LY-CoV016 and REGN10933, little effect on REGN1097 or LY-CoV555
L452: mutations reduce binding by LY-CoV555, little effect on REGN10933, REGN01987, LY-CoV016. (6/n)
Importantly, these maps also have data for all other possible mutations, so as new mutations appear you can check if therapeutic antibodies will be impacted. Important to monitor this, since some of the antibodies are affected by mutations already appearing. (7/n)
Two other main findings in the paper. First, it describes how viral antibody-escape mutations arose in persistently infected patient treated w REGN-COV2. Since we posted pre-print, other studies have also reported viral antigenic evolution in persistently infected patients. (8/n)
Second, describes single amino-acid mutation E406W that escapes both antibodies in REGN-COV2. We don't think E406W is clinical risk as requires multiple nucleotide changes to codon, but would be interesting to know biochemical mechanism (we hope others are working on this!) (9/n)
Finally, all of this work including the unpublished LY-CoV555 data above was led by @tylernstarr, with important contributions from @AllieGreaney, @AdamDingens, @Dr_MChoudhary, @DrJLi, Amin Addetia, and Will Hannon. (10/n)
• • •
Missing some Tweet in this thread? You can try to
force a refresh