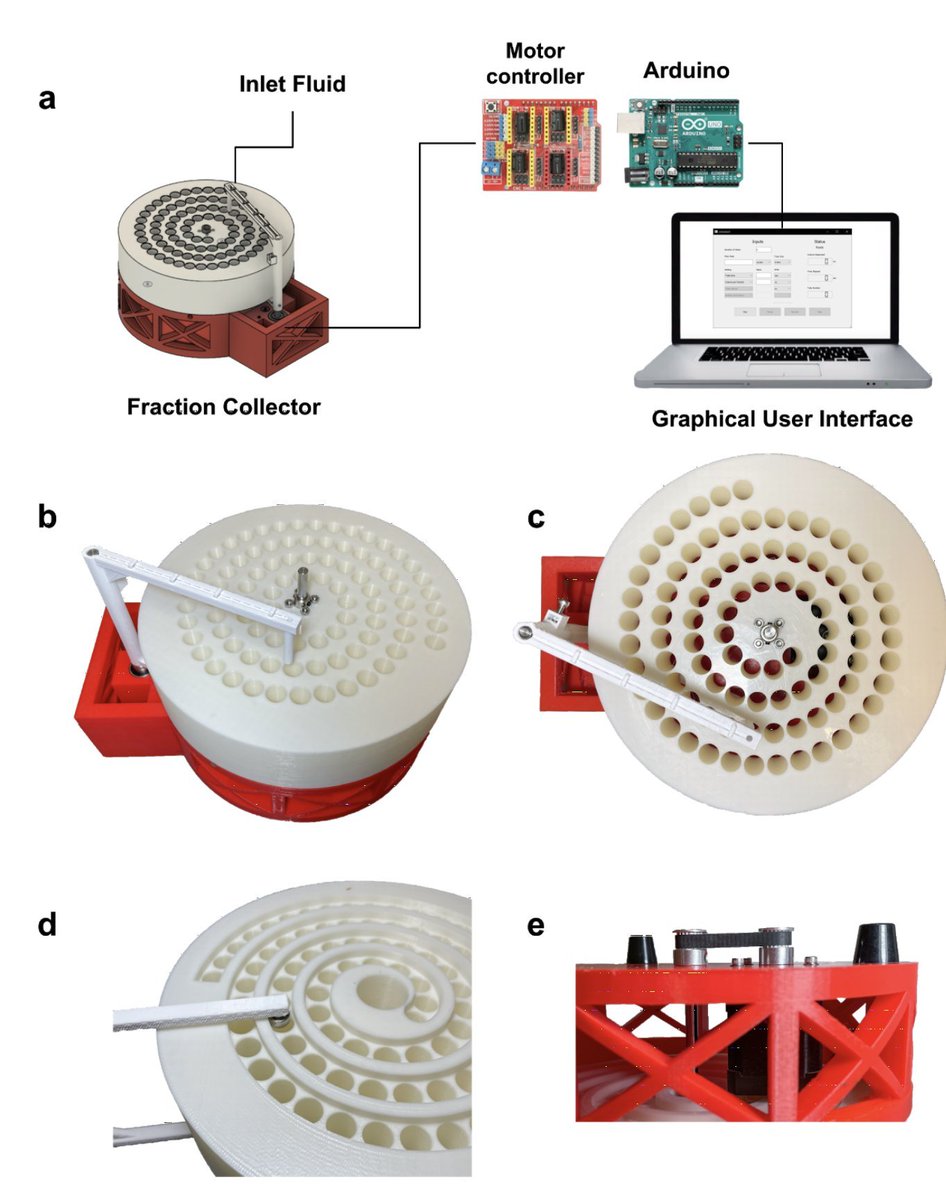
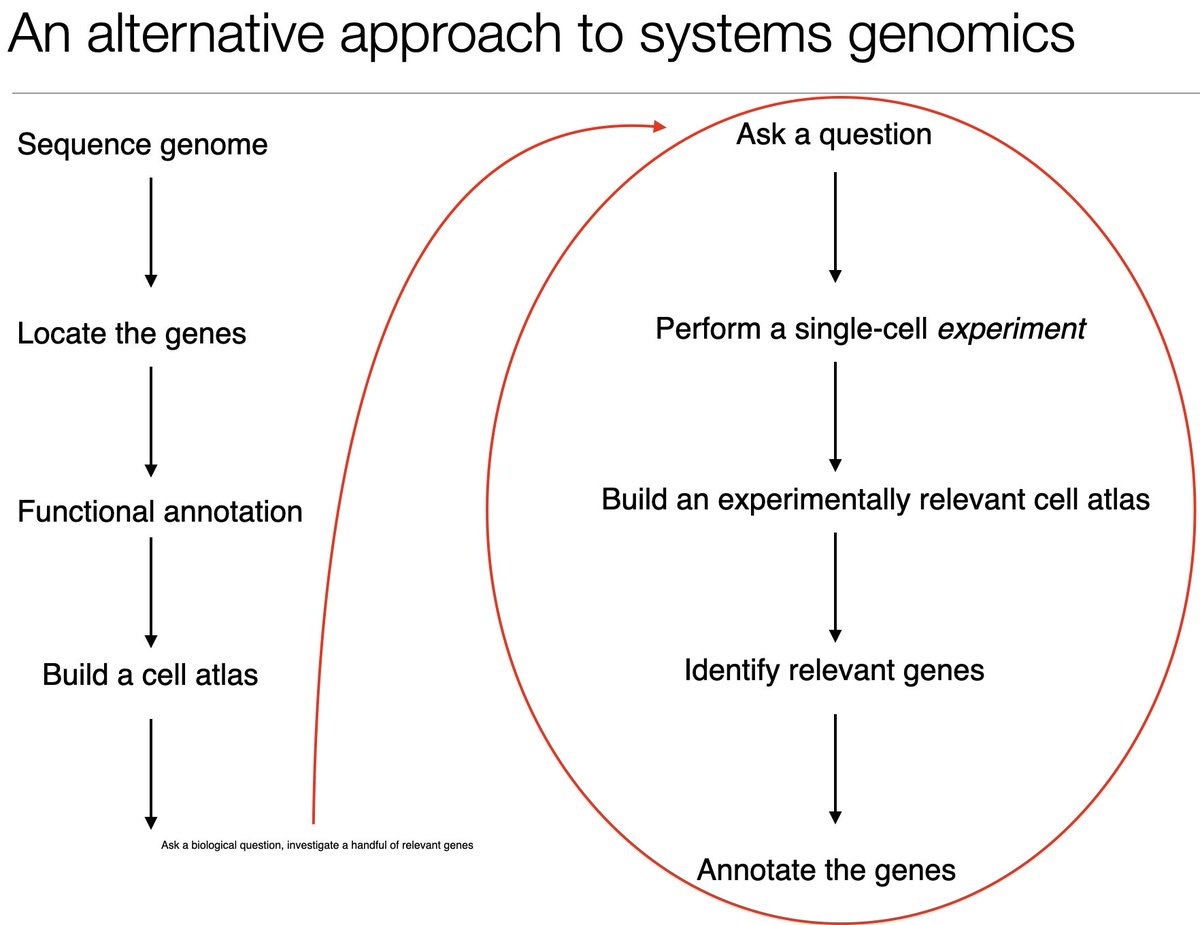
"The new answer to a 77-year-old problem"
😭
😭
https://twitter.com/adamcchang/status/1141383113987117056
You've come this far and you're thinking... oh... but plot the difference...
Ok. biorxiv.org/content/10.110…
Ok. biorxiv.org/content/10.110…
I will say, box plots / dynamite plots are a terrible way to display data and too many people still use them. But people make a lot of poor choices in their figures. Since you've come this far I'll share this ultimate useless figure... enjoy Figure 1... jstor.org/stable/2291148…
• • •
Missing some Tweet in this thread? You can try to
force a refresh