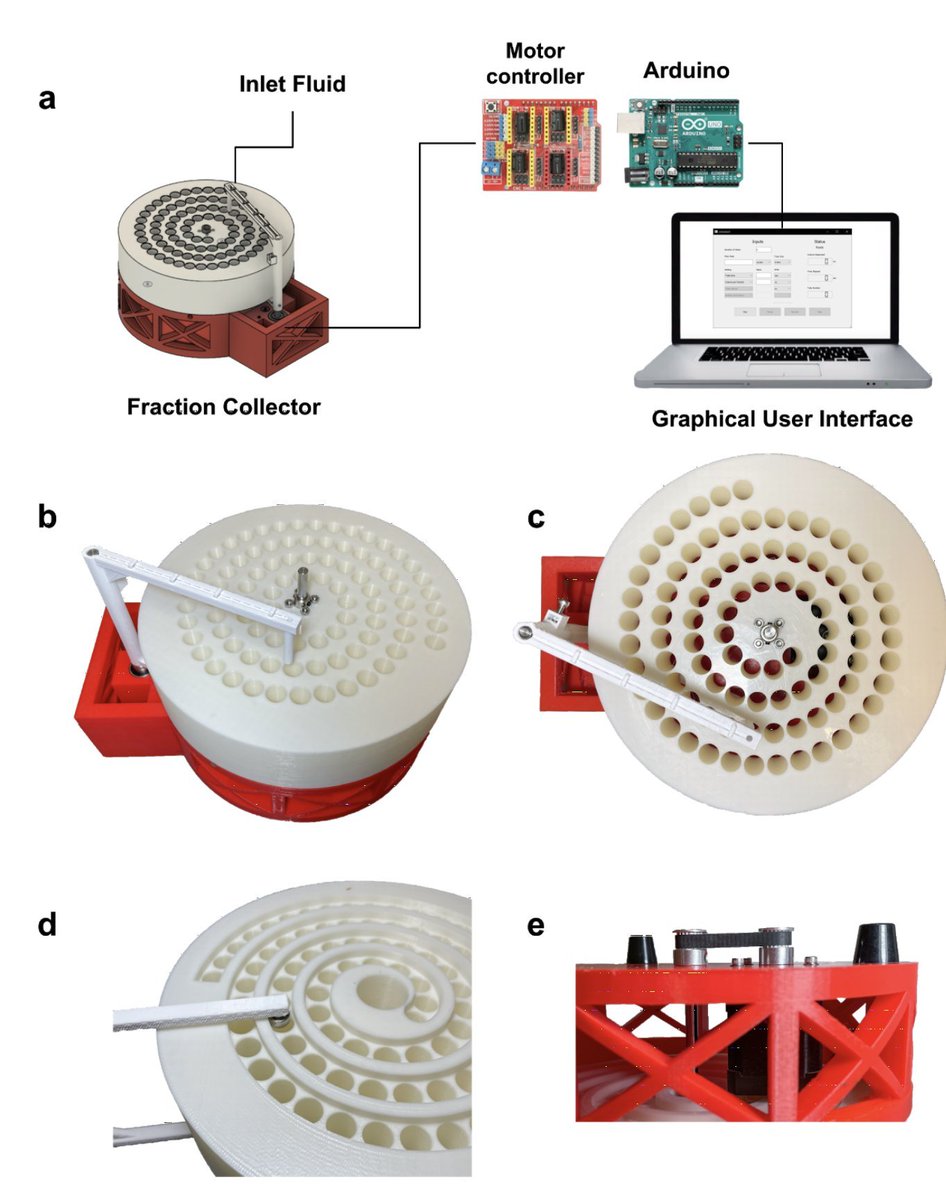
Happy to announce a new open-source instrument from our lab, this time a low-cost, scalable, and automated fraction collector for fluidics applications. Beautiful work by @sinabooeshaghi & @annekylosaurus, with @lioscro and @JaseGehring. 1/
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
The design is simple and elegant. A single motor drives the shaft of the tube rack, which is coupled to the dispenser arm via a spiral track. This ensures both rotate in tandem. 2/ 

The device is easy to 3D print and build, and can be assembled from off-the-shelf parts in less than an hour for $67.02. This low cost, and the straightforward assembly, is possible thanks to the design around a single motor. Amazing work by @annekylosaurus & @sinabooeshaghi. 3/
The colosseum fraction collector is run via a pip installable GUI thanks to @lioscro. The easy installation is matched by ease of use. Detailed instructions are posted on the project Github repository: github.com/pachterlab/col… 4/ 

For more details on building and running the instrument, see @annekylosaurus' instructional video: 5/
The preprint includes careful benchmarks of the instrument. These were produced using flow produced with our open source poseidon syringe pump system. github.com/pachterlab/pos… 6/ 

There are some interesting math questions that arise in this application, as related to the use of an Archimedean spiral. We compared our packing efficiency to the optimal packing efficiency and found we were at 62%. We will try to improve on this in a future release. 7/ 

The benchmarking results, and other calculations, are all fully reproducible via notebooks in a Github repository that accompanies the paper. There are [code] links next to each figure linking the notebooks. github.com/pachterlab/BKM… 8/
One of the great things about our fraction collector, is that unlike commercial devices, it is easily customizable, both in software and in hardware. We've shared all the STL files and all the source code under a BSD-2 open source license. Want different size🧪? No problem. 9/
This project is a sequel to the poseidon syringe pump project (Booeshaghi et al., 2019 nature.com/articles/s4159…). We're been happy to see it widely adopted, for projects ranging from Drop-seq to high-school education kits. 10/
We've been particularly thrilled to see this "translation research" (bench to school) possible thanks to poseidon: pubs.acs.org/doi/abs/10.102… Hopefully the colosseum fraction collector opens the door to many more applications and experiments. 11/
Special thanks to @tdilan4 who helped with the experiments & benchmarking. Thanks also to @VeigaBeltrame for help with 3D printing. Finally, thanks to the users of the poseidon pumps who have given us valuable feedback that has helped shaped this project. More to come! 12/12.
• • •
Missing some Tweet in this thread? You can try to
force a refresh