Our complete mapping of mutations to #SARSCoV2 RBD that reduce binding by convalescent human plasma is out in @cellhostmicrobe (cell.com/cell-host-micr…). Right now E484K getting lot of attention, but I want to emphasize what our results suggest to keep eyes on in *future* (1/n)
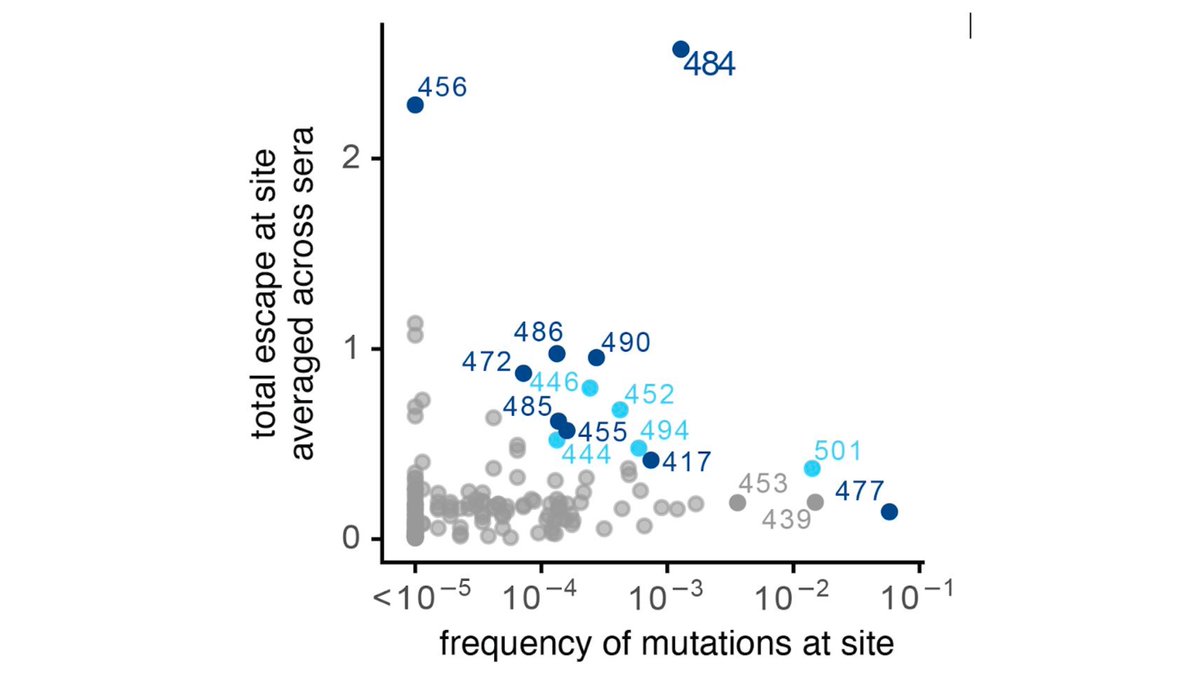
To recap, we measured how all mutations to RBD reduce binding by antibodies in convalescent plasma. Lots of person-to-person variation in effects of mutations, but mutations at E484 have biggest effect. My old summary from early Jan:
https://twitter.com/jbloom_lab/status/1346442000472580098(2/n)
That summary was written just as E484K-containing 501Y.V2 (B.1.351) & 501Y.V3 (P.1) lineages were being reported & focused on E484 as most important site of mutations. Since then, many labs have characterized these lineages to confirm E484K is major antigenic change. (3/n)
But our approach doesn't involve reactively characterizing mutations after they are observed in new lineages: rather, it prospectively maps effects of all mutations so we can know what to watch for next. So what to watch for? (4/n)
Well, E484 mutations are single worst change, & are already with us. But our maps show that next most important epitope for polyclonal convalescent antibodies is 443-450 loop in RBD (which peripherally includes sites like 452 & 494; see paper &
https://twitter.com/jbloom_lab/status/1346442010132054017). (5/n)
Indeed, for minority of people, mutations in 443-450 loop (eg, at G446 as below
https://twitter.com/jbloom_lab/status/1346442020051537922) already have bigger effect than E484K. And if E484 epitope is ablated as E484K spreads, I'd expect 443-450 loop to take on growing importance for remaining immunity. (6/n)
More generally, our maps measure effects of all mutations, so hope people refer back to them as new mutations are observed.
Finally, all of above results come from great efforts of @AllieGreaney, Andrea Loes, @khdcrawford, @tylernstarr, Keara Malone, @HelenChuMD. (7/n)
Finally, all of above results come from great efforts of @AllieGreaney, Andrea Loes, @khdcrawford, @tylernstarr, Keara Malone, @HelenChuMD. (7/n)
• • •
Missing some Tweet in this thread? You can try to
force a refresh