New SARS-CoV-2 variant update for Connecticut.
- 42 cases of B.1.1.7
- 1 case of B.1.351
- 5 cases of B.1.525 (not of concern yet, but we are monitoring)
Partnership with @jacksonlab, @CTDPH, and diagnostic labs
Follow the 🧵(1/10)
Or read the report 👇
covidtrackerct.com/variant-survei…
- 42 cases of B.1.1.7
- 1 case of B.1.351
- 5 cases of B.1.525 (not of concern yet, but we are monitoring)
Partnership with @jacksonlab, @CTDPH, and diagnostic labs
Follow the 🧵(1/10)
Or read the report 👇
covidtrackerct.com/variant-survei…
2/10
B.1.1.7 cases in Connecticut by county.
- New Haven County: 33
- Fairfield County: 3
- Litchfield County: 3
- Hartford County: 2
- Windham County: 1
We do most of our surveillance from New Haven County, hence the higher numbers. We'll have a map next week.
B.1.1.7 cases in Connecticut by county.
- New Haven County: 33
- Fairfield County: 3
- Litchfield County: 3
- Hartford County: 2
- Windham County: 1
We do most of our surveillance from New Haven County, hence the higher numbers. We'll have a map next week.
3/10
B.1.1.7 cases in Connecticut are on the rise, but we don't yet know its frequency in the community. Its likely still <3% based on the PCR data.
B.1.1.7 cases in Connecticut are on the rise, but we don't yet know its frequency in the community. Its likely still <3% based on the PCR data.

4/10
We have evidence for multiple introductions of B.1.1.7 into CT (left figure), and evidence for community transmission (right figure, identical genomes).

We have evidence for multiple introductions of B.1.1.7 into CT (left figure), and evidence for community transmission (right figure, identical genomes).


5/10
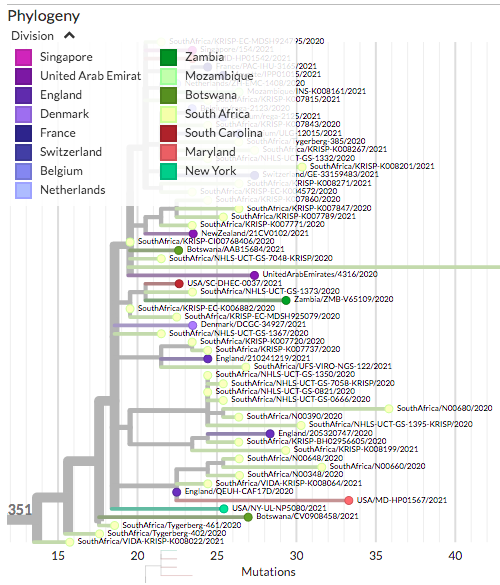
The first case of B.1.351 in CT was announced yesterday (detected in NY, CT resident). Its unrelated to the other B.1.351 sequences from MD and SC.
USA/NY-UL-NP5080/2021
nextstrain.org/community/grub…
The first case of B.1.351 in CT was announced yesterday (detected in NY, CT resident). Its unrelated to the other B.1.351 sequences from MD and SC.
USA/NY-UL-NP5080/2021
nextstrain.org/community/grub…
6/10
P.1 has not yet been detected in Connecticut, though it is possible that it has been introduced and is circulating.
P.1 has not yet been detected in Connecticut, though it is possible that it has been introduced and is circulating.
7/10
B.1.525 is a new variant that we are monitoring. It has several key mutations: E484K, Q677H, F888L and a similar suite of deletions to B.1.1.7. We've detected 5 cases with this variant in CT. We don't yet know its impact on vaccines.
nextstrain.org/community/grub…
B.1.525 is a new variant that we are monitoring. It has several key mutations: E484K, Q677H, F888L and a similar suite of deletions to B.1.1.7. We've detected 5 cases with this variant in CT. We don't yet know its impact on vaccines.
nextstrain.org/community/grub…

8/10
We are also monitoring a lineage of viruses that have a 501T mutation. These are found throughout CT. Significance is unknown.

We are also monitoring a lineage of viruses that have a 501T mutation. These are found throughout CT. Significance is unknown.


9/10
Led by @tdalpert, we recently posted a preprint describing the establishment of B.1.1.7 across the US and the urgent need to enhance surveillance.
Read 👇
medrxiv.org/content/10.110…
Led by @tdalpert, we recently posted a preprint describing the establishment of B.1.1.7 across the US and the urgent need to enhance surveillance.
Read 👇
medrxiv.org/content/10.110…

10/10
Our variant reports will be updated once a week on Tuesdays. Huge thanks to our team that unselfishly keeps this going, every week. @JosephFauver, @tdalpert, @MaryPetrone10, @VogelsChantal, Mallery Breban, @aewatkins6, @AndersonBrito_, @ChaneyKalinich, @RebeccaEarnest1
Our variant reports will be updated once a week on Tuesdays. Huge thanks to our team that unselfishly keeps this going, every week. @JosephFauver, @tdalpert, @MaryPetrone10, @VogelsChantal, Mallery Breban, @aewatkins6, @AndersonBrito_, @ChaneyKalinich, @RebeccaEarnest1
• • •
Missing some Tweet in this thread? You can try to
force a refresh