In new work led by @AllieGreaney, we analyze mutational escape of #SARSCoV2 from monoclonal & polyclonal antibodies in terms of RBD epitope classes (biorxiv.org/content/10.110…). Provides useful framework for conceptualizing effects of individual and combined mutations. (1/n)
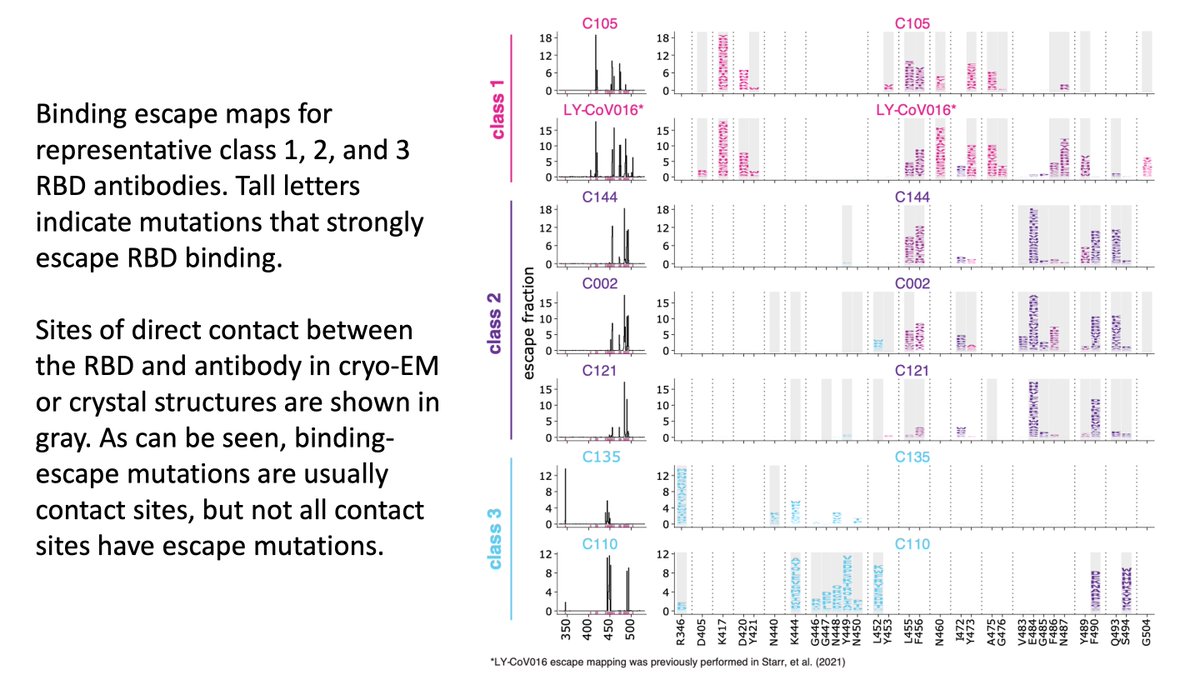
Specifically, @cobarnes27 @bjorkmanlab classified potent neutralizing anti-RBD antibodies in 3 classes using structural analyses (nature.com/articles/s4158…). These classes (1, 2, 3) shown below (also 4th class of less potent antibodies that bind further from ACE2 interface). (2/n) 

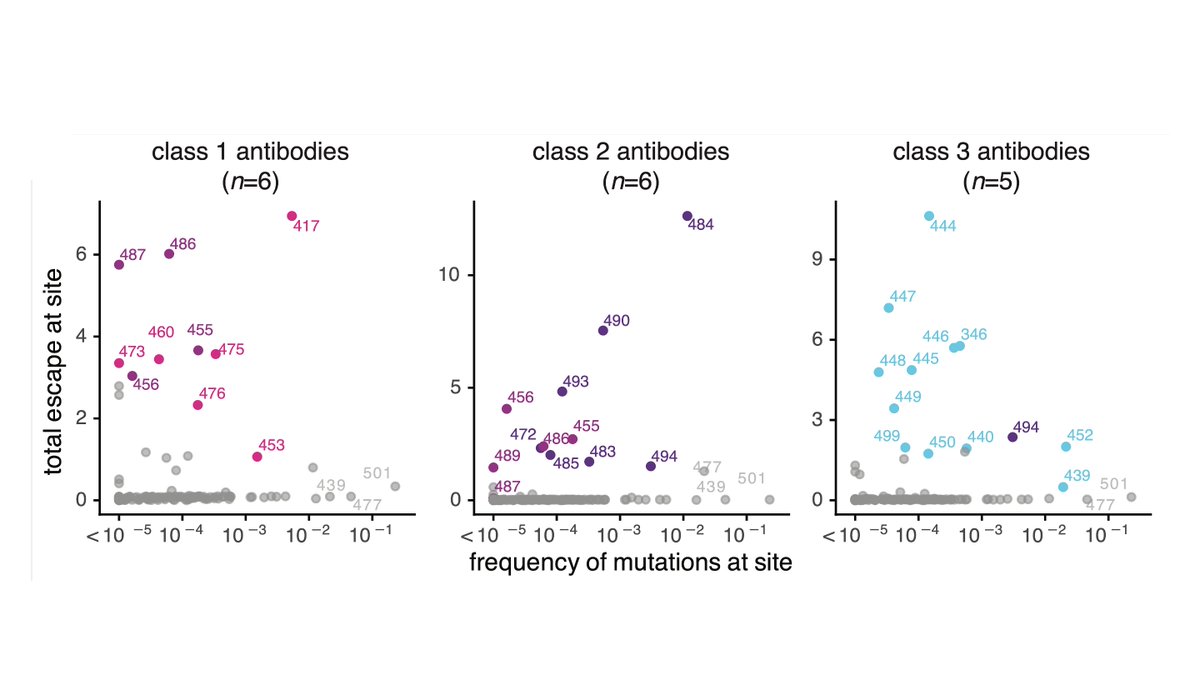
@AllieGreaney used deep mutational scanning to map all mutations that escape binding to yeast-displayed RBD by antibodies of each class (from @NussenzweigL). Below are escape maps. Escape mutations usually at antibody contact sites, but not all contact site mutations escape (3/n) 
We performed multi-dimensional scanning on the binding-escape maps to organize antibodies in the space of “viral escape.” Antibodies nearby in this space are escaped by similar mutations. (4/n) 

We then mapped escape from polyclonal plasma, including for individuals from whom some of the antibodies isolated. Mutations that reduce plasma binding most similar class 2 antibody escape, even for individuals from whom antibodies of other classes were isolated. (5/n) 
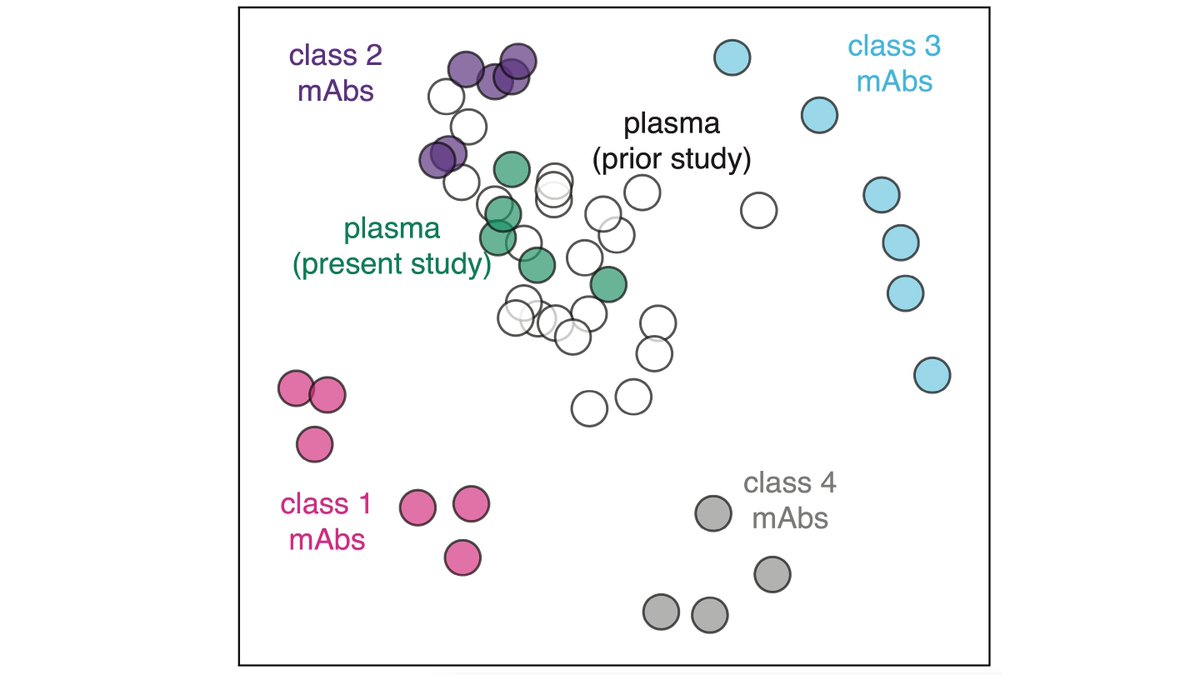
Therefore, even though immune system can make many different types of antibodies, a single class of antibodies usually functionally dominates the anti-RBD portion of the polyclonal response to infection. (6/n)
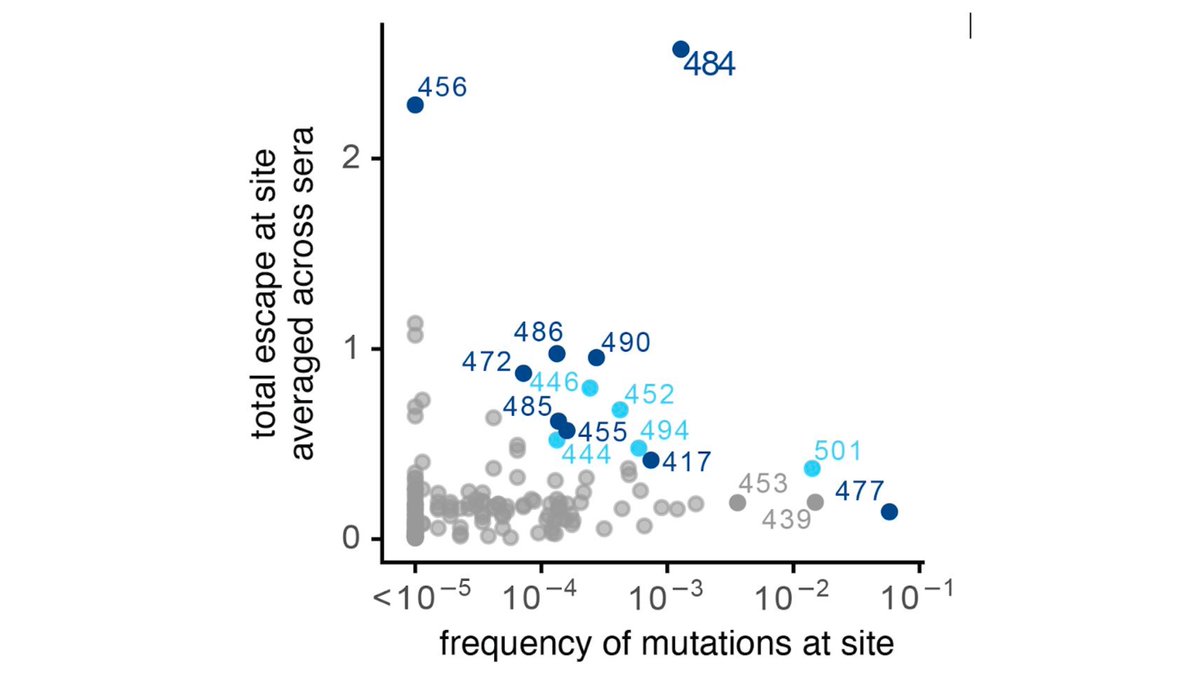
We analyzed which antibody classes escaped by emerging #SARSCoV2 mutations. Mutations spreading at sites of biggest escape for class 1 (K417) & class 2 (E484). But mutations at major class 3 escape sites (K444, G446, G447) not common, although L452 is modest class 3 escape (7/n) 
Can also look in context of viral lineages (see table). Several lineages (B.1.351, P.1) combine escape mutations for two antibody classes, but none escape all three.
*Emergence of a major class 3 escape mutation in background of class 1 + 2 escape would be concerning.* (8/n)
*Emergence of a major class 3 escape mutation in background of class 1 + 2 escape would be concerning.* (8/n)

Note caveat that we are only looking at RBD mutations here; NTD antibodies have distinct epitopes, so a full analysis should look for escape from different classes of RBD antibodies plus escape from NTD antibodies. (9/n)
Cool aside: @PaulBieniasz @theodora_nyc have performed elegant VSV-spike escape selections with many of these antibodies. If we overlap @AllieGreaney’s binding escape & @tstarr’s ACE2-affinity deep mutational scanning, great concordance with their viral selections! (10/n) 

Finally, enjoyable part of project was getting to build on nice prior work by @cobarnes27 @bjorkmanlab @NussenzweigL @PaulBieniasz @theodora_nyc. We’ve read & enjoyed their papers on #SARSCoV2 antibodies, so appreciated opportunity to get to extend some parts of it. (11/n)
• • •
Missing some Tweet in this thread? You can try to
force a refresh