B.1.618 - a new lineage of SARS-CoV-2 predominnatly found in India and characterized by a distinct set of genetic variants including E484K , a major immune escape variant. 
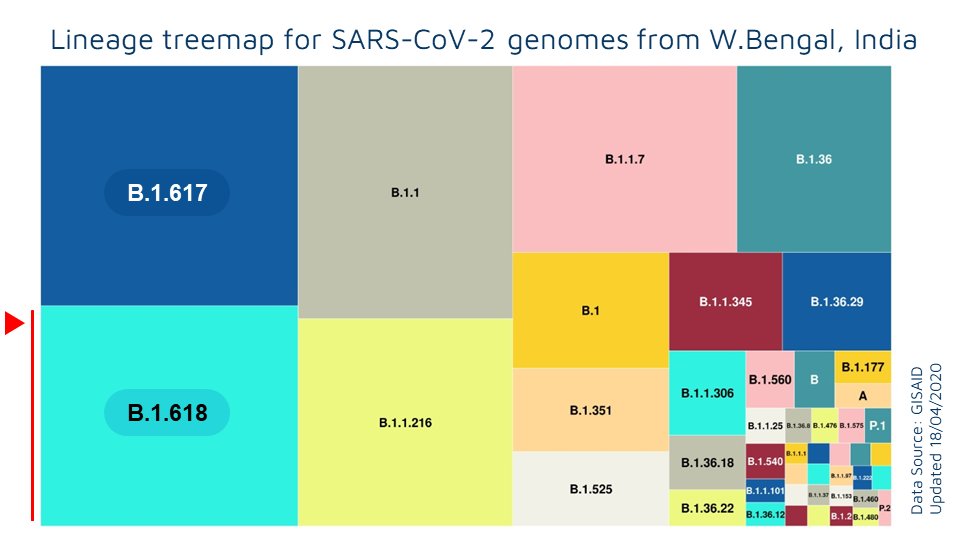
Initial sequences in the B.1.618 lineage were found in West Bengal, India.
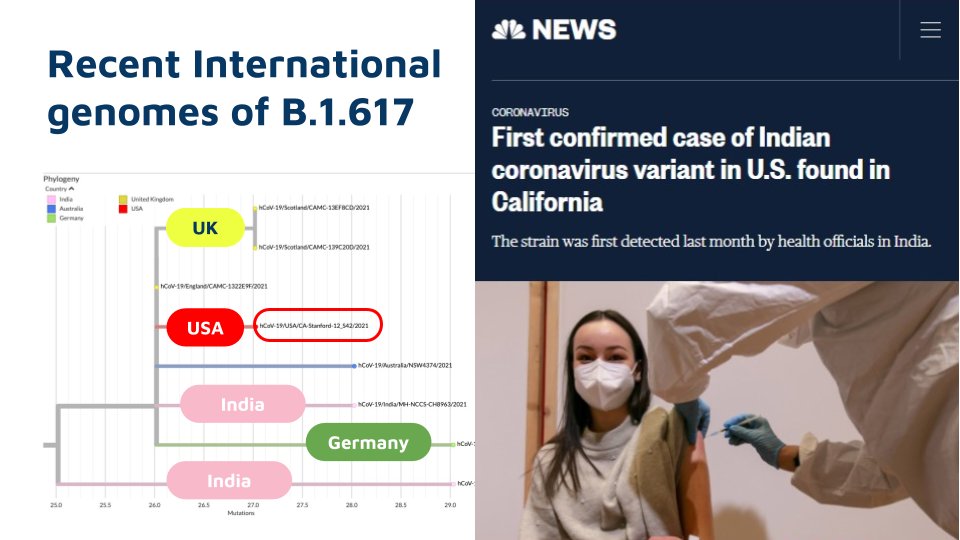
Members of this lineage is also found in other parts of the world, cov-lineages.org/lineages/linea… but do not have the full complement of variants as found in India
Members of this lineage is also found in other parts of the world, cov-lineages.org/lineages/linea… but do not have the full complement of variants as found in India
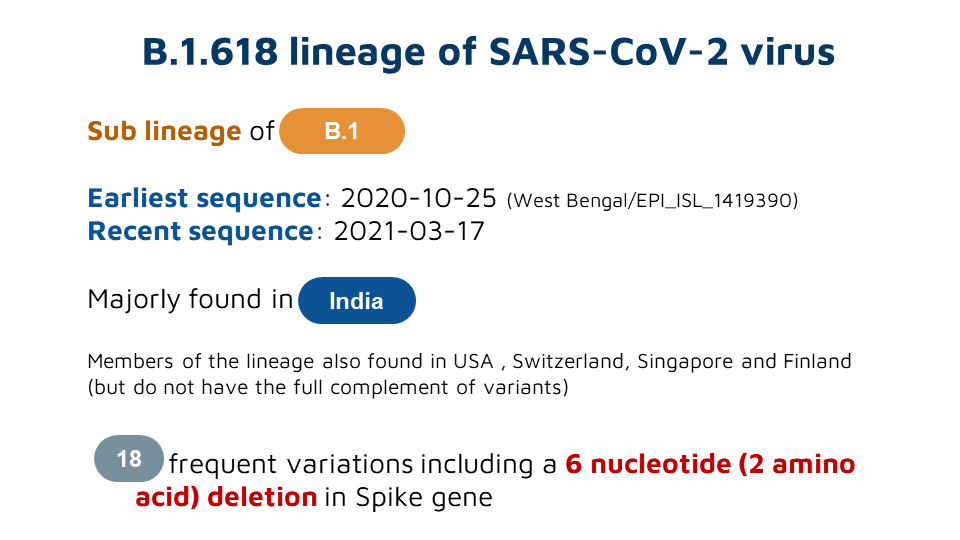
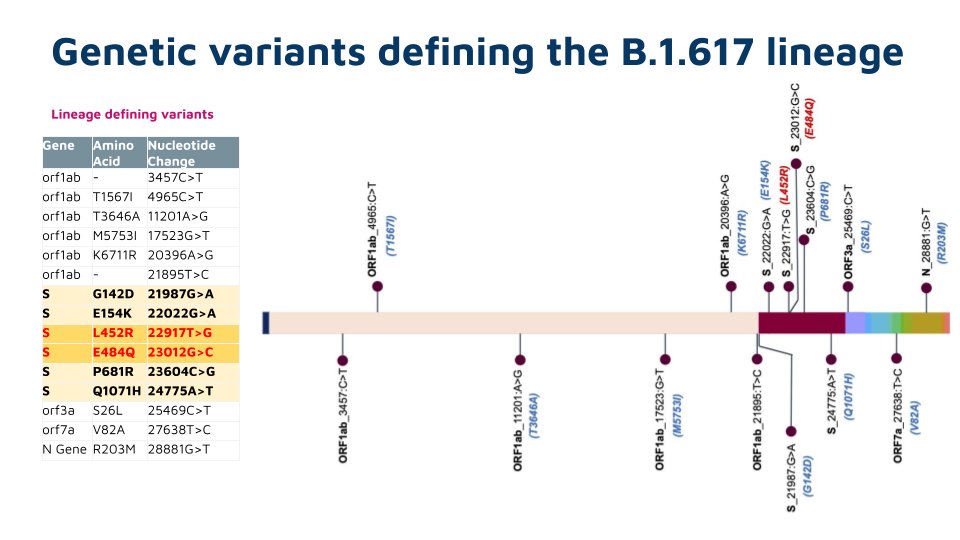
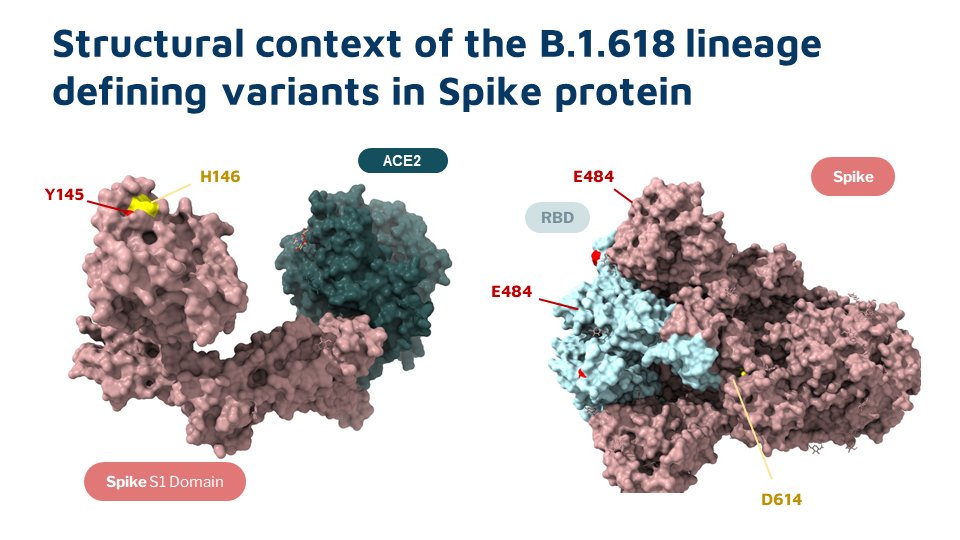
This lineage is characterized by a 6 nt deletion (H146del &Y145del) , apart from E484K and D614G in spike protein
Other variants are in the ORF1ab, ORF3a, ORF7a, ORF7b and N genes
Other variants are in the ORF1ab, ORF3a, ORF7a, ORF7b and N genes
E484K is a major immune escape variant - also found in a number of emerging lineages across the world.
E484K can escape multiple mAbs as well as panels of convalescent plasma.
The current evidence available on immune escape variants are compiled at: clingen.igib.res.in/esc/
E484K can escape multiple mAbs as well as panels of convalescent plasma.
The current evidence available on immune escape variants are compiled at: clingen.igib.res.in/esc/
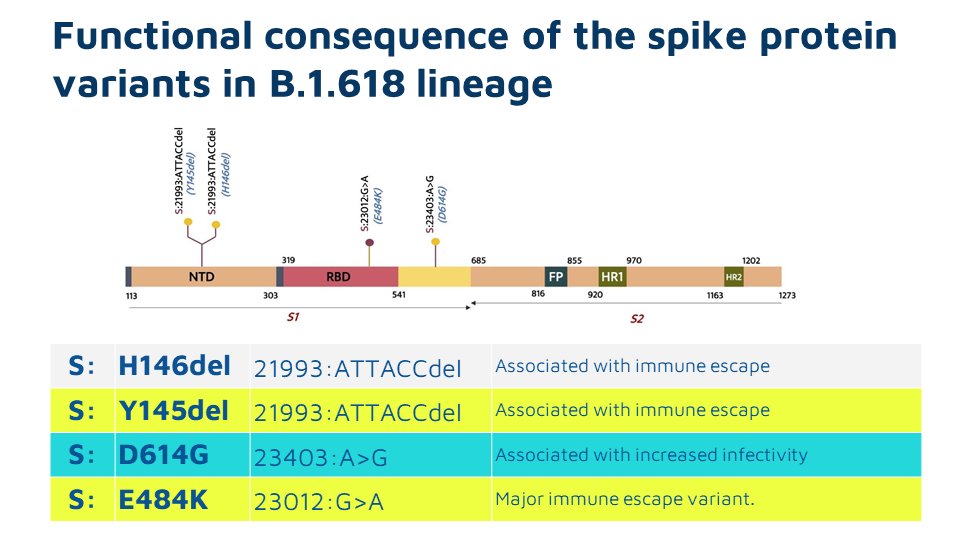
While E484K is in the Receptor Binding Domain, Y145 and H146 are not part of the residues interacting with the Human ACE2 receptor. The structural impact of the 2AA deletion causes to spike protein is yet to be understood completely. 
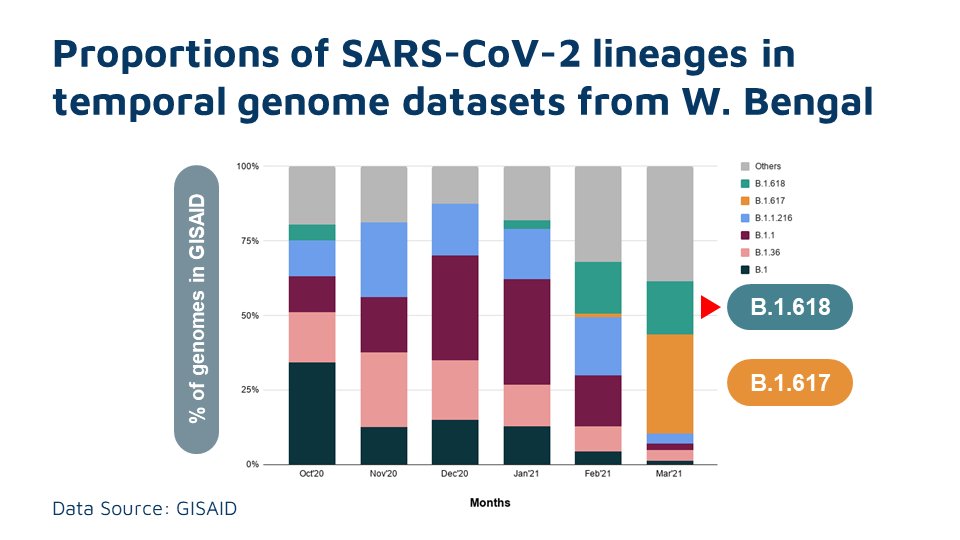
The proportions of B.1.618 has been growing significantly in the recent months in the state of West Bengal, India 
There are many unknowns for this lineage at this moment including its capability to cause reinfections as well as vaccine breakthrough infections. Additional experimental data is also required to assess the efficacy of vaccines against this variant.
At this moment, there is no conclusive evidence that the lineage drives the epidemic in West Bengal, apart from the fact that the nos and proportions have been significantly increasing in recent months. More focused epidemiological investigations would address these questions.
• • •
Missing some Tweet in this thread? You can try to
force a refresh