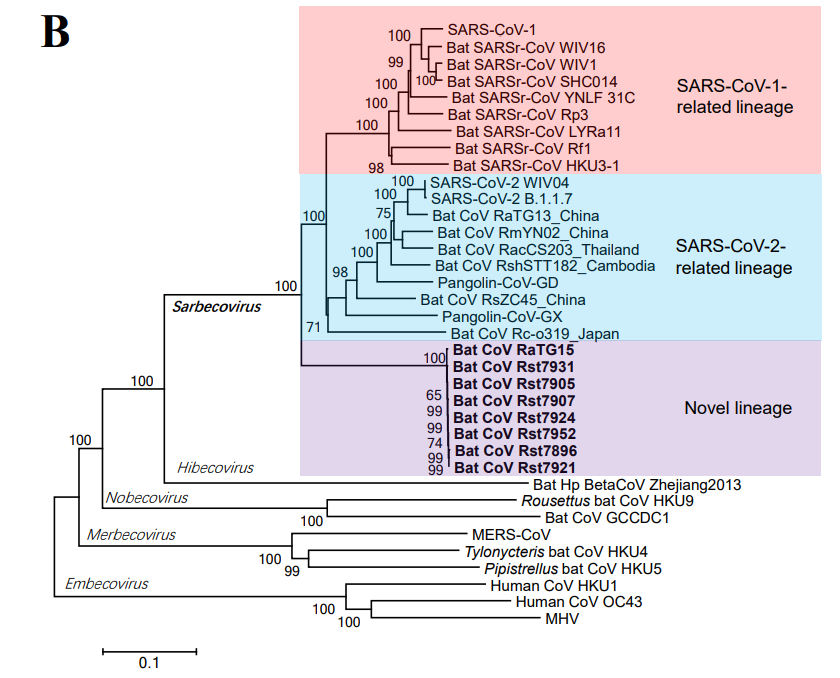
They finally concede after more than a year: "Here, we report the identification of a novel lineage of SARSr-CoVs, including RaTG15 and seven other viruses, from bats at the same location where we found RaTG13 in 2015"
No mention to the miners or the mine. Just this: "in Tongguan town, Mojiang county, Yunnan province in China in 2015, the same location where we found bat
RaTG13 in 2013"
RaTG13 in 2013"
Yes, the same 8 viruses that were embargoed from November 2019 to June 2020.
Gentle reminder:
Gentle reminder:
https://twitter.com/franciscodeasis/status/1366750802057064449
Attention to this. They deposited the sequences only in a Chinese repository. They have learned it is more difficult to censor critical data if deposited in foreign repositories. 

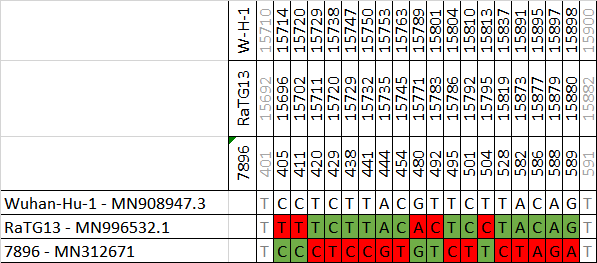
No surprises, the RBD aa seq of Ra7909 is the same as in that slide that disappeared from the webinars.
https://twitter.com/franciscodeasis/status/1366823019755294723
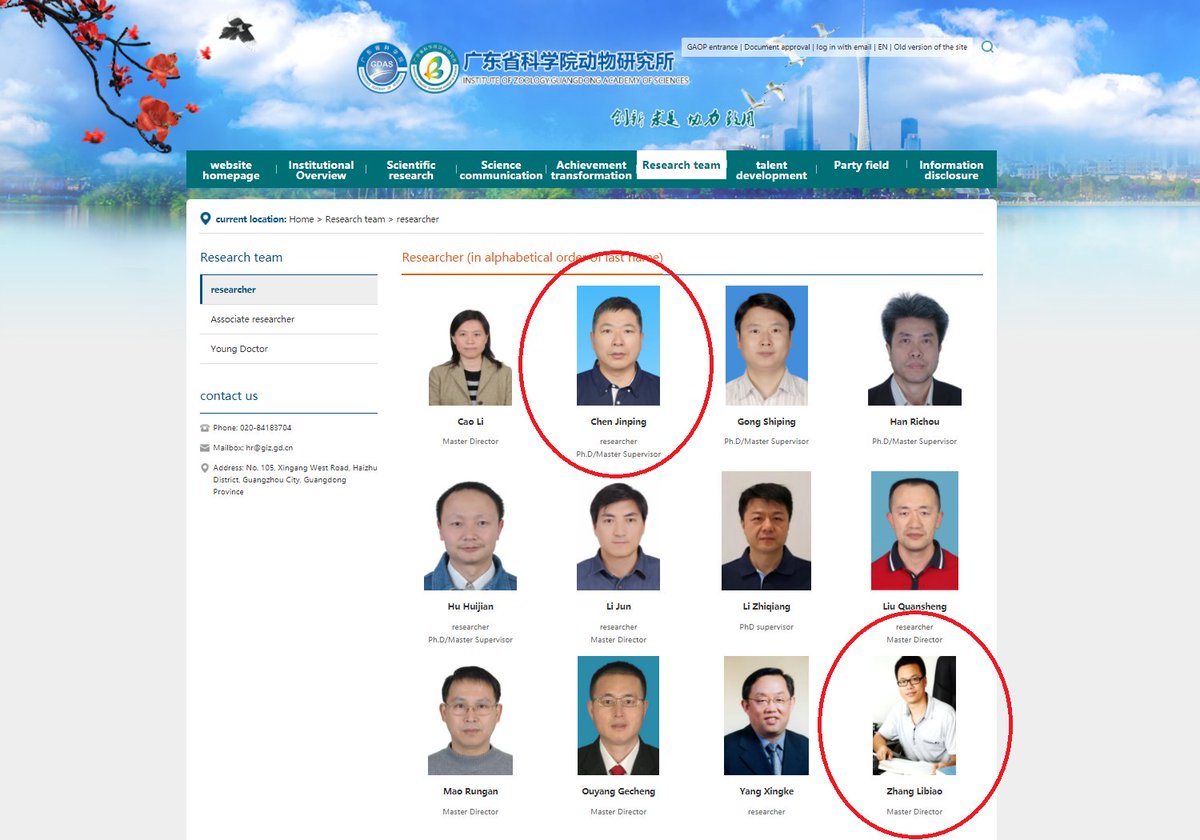
Now it is clear who took the samples. One of the three we already tried to guess: XLY

https://twitter.com/franciscodeasis/status/1333543865614217216

"small" lapsus. The pre-print is focused on RaTG13 (or Ra7909, although association is not explicit). All the other viruses only appear one time: in Fig S1B. But "7896" seems special for naming FTP folders. If they had used a US repository, this would not have happened to them 

We now know the exact day of the trip. Same day for the range 7896-7952:
2015-05-29


2015-05-29
https://twitter.com/franciscodeasis/status/1332700756185636866


Gentle reminder on how they tried to hide it before:
https://twitter.com/franciscodeasis/status/1347959947036942336
The low variability we observed among the 8 RdRps has now been reduced more.
Old 7952 RdRp (MN312673) vs new 7952 RdRp (GWHBAUT01000001).
Changes exactly where 7952 was different vs the other viruses.
Now 7952 and 7909 RdRps are the same

Old 7952 RdRp (MN312673) vs new 7952 RdRp (GWHBAUT01000001).
Changes exactly where 7952 was different vs the other viruses.
Now 7952 and 7909 RdRps are the same
https://twitter.com/franciscodeasis/status/1372581427502592007

7952 and 7909 are now only 8 nt different (in full genome!). This is the most strange thing for me. In Jinning the variability was huge. It was even mentioned in the new discovered theses
https://twitter.com/franciscodeasis/status/1371616712081162242
And remember that 7952 seems to have an unpublished co-infection. What if it happened the same as in Jinning?
https://twitter.com/franciscodeasis/status/1371509838883209216
That would be Longtanxiang not Tongguanzhen
Correct location of the mine:
23°10'44.9"N 101°22'04.3"E
cc @TheSeeker268 @Drinkwater5Reed & #DRASTIC
23°10'44.9"N 101°22'04.3"E
cc @TheSeeker268 @Drinkwater5Reed & #DRASTIC
https://twitter.com/franciscodeasis/status/1370527305680949249
Let's make a bet, what name do you think they will choose again for next week?
Do not miss these tools of @babarlelephant. Babar was the 1st to show the proximity of these 8 viruses to SARS-CoV-2 in a phylogenetic tree.
AFAIK that tree was the only existing tree with these 8 viruses for 6 months.
Note: you can change "query" to 7909
AFAIK that tree was the only existing tree with these 8 viruses for 6 months.
Note: you can change "query" to 7909
https://twitter.com/babarlelephant/status/1395935610951581696
• • •
Missing some Tweet in this thread? You can try to
force a refresh