Excited to introduce #3DRCAN for denoising, super resolution and expansion microscopy. Super proud of this colab with Hari Shroff, Jiji Chen et al.
@NIBIBgov @arpcomplex
Preprint: biorxiv.org/content/10.110…
Code: github.com/AiviaCommunity…
#microscopy #aimicroscopy #aivia
1/17
@NIBIBgov @arpcomplex
Preprint: biorxiv.org/content/10.110…
Code: github.com/AiviaCommunity…
#microscopy #aimicroscopy #aivia
1/17
"Three-dimensional residual channel attention networks denoise and sharpen fluorescence microscopy image volumes"
J Chen, H Sasaki, H Lai, Y Su, J Liu, Y Wu, A Zhovmer, C Combs, I Rey-Suarez, H Chang, C Huang, X Li, M Guo, S Nizambad, A Upadhyaya, J Lee, L Lucas, H Shroff.
2/17
J Chen, H Sasaki, H Lai, Y Su, J Liu, Y Wu, A Zhovmer, C Combs, I Rey-Suarez, H Chang, C Huang, X Li, M Guo, S Nizambad, A Upadhyaya, J Lee, L Lucas, H Shroff.
2/17
We are particularly interested in characterizing the limits of #deeplearning based approaches to image restoration. You will find several experiments comparing several best-in-class approaches: #3DRCAN, #CARE, #SRResNet and #ESRGAN.
#microscopy #aimicroscopy #iSIM
3/17
#microscopy #aimicroscopy #iSIM
3/17
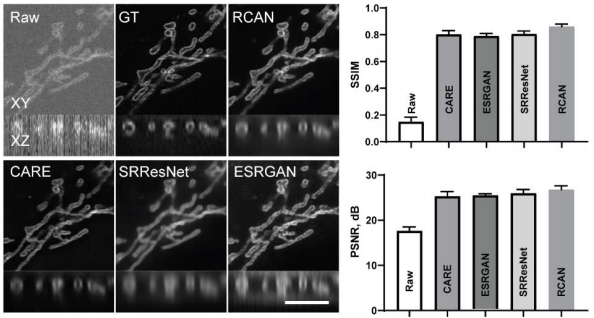
Firstly, we looked at how #3DRCAN and other #DL based approaches behave when denoising iSIM images depicting a range of organelles, including: Actin, ER, Golgi, Lysosome, Microtubule, Mitochondria.
#microscopy #aimicroscopy #cellbio #deeplearning #superresolution
4/17
#microscopy #aimicroscopy #cellbio #deeplearning #superresolution
4/17
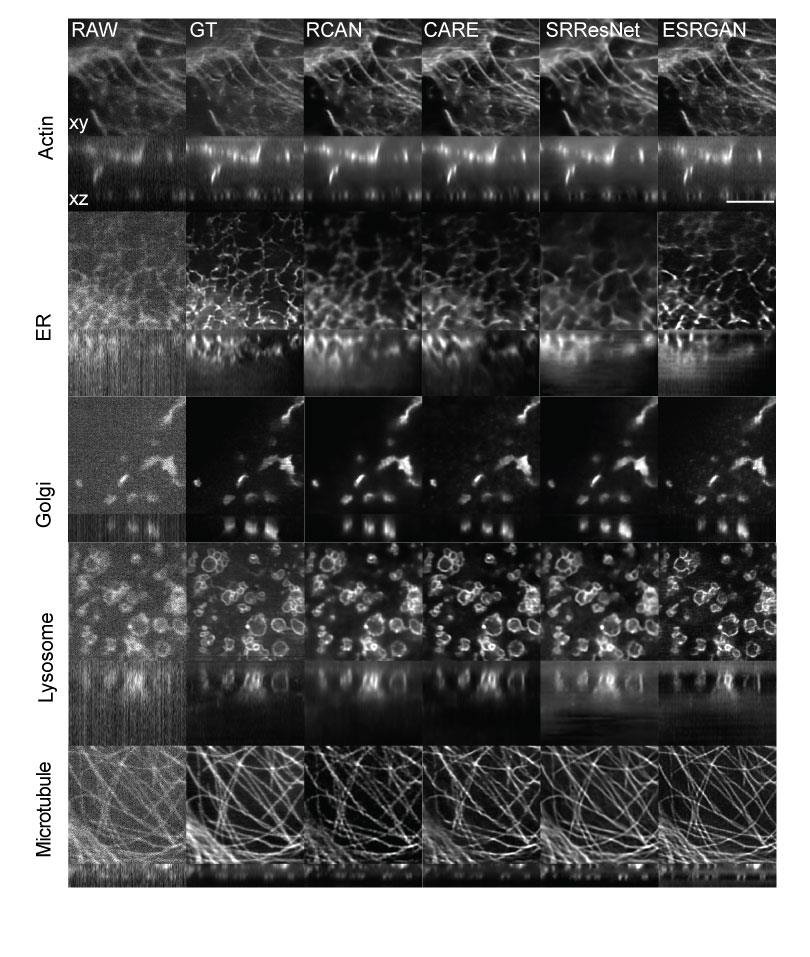
We were also interested in finding out the lowest level of illumination (and lowest SNR) for which #3DRCAN could still "work", i.e. create a good quality restoration. Imaging done on an iSIM.
#iSIM #microscopy #aimicroscopy #cellbio #deeplearning #bioimageanalysis #aivia
5/17
#iSIM #microscopy #aimicroscopy #cellbio #deeplearning #bioimageanalysis #aivia
5/17
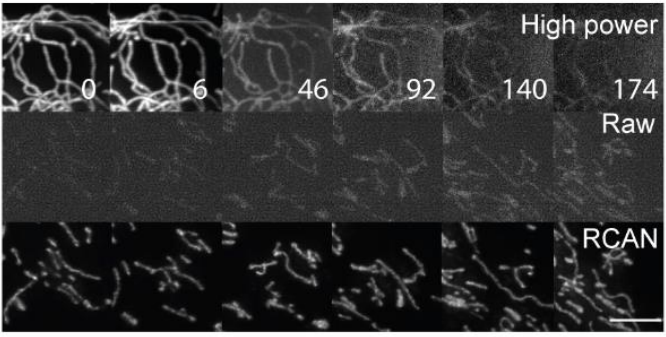
Now that we had nicely restored images (good performance in terms of SSIM and PSNR), we detected and analyzed mitochondria. This was nearly impossible when using the raw/input data.
#microscopy #aimicroscopy #cellbio #deeplearning #bioimageanalysis #aivia #3DRCAN
6/17


#microscopy #aimicroscopy #cellbio #deeplearning #bioimageanalysis #aivia #3DRCAN
6/17



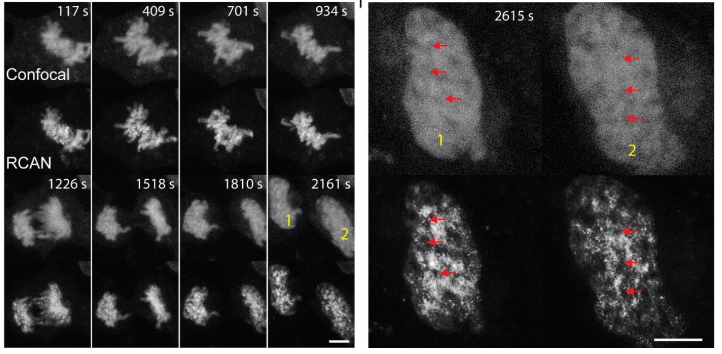
We then applied the trained #3DRCAN to live cell imaging data (4D). As a result we could study fusion and fission (as imaged by iSIM) events in mitochondria over long time lapses.
#microscopy #aimicroscopy #cellbio #deeplearning #bioimageanalysis
7/17

#microscopy #aimicroscopy #cellbio #deeplearning #bioimageanalysis
7/17


Secondly, we wanted to determine if #3DRCAN could be used for other types of super resolution microscopy - we used paired Confocal<->STED 3D data sets to train our models. Imaging done with a Leica SP8. @LeicaMicro
#STED #superresolution #aimicroscopy #microscopy
8/17
#STED #superresolution #aimicroscopy #microscopy
8/17
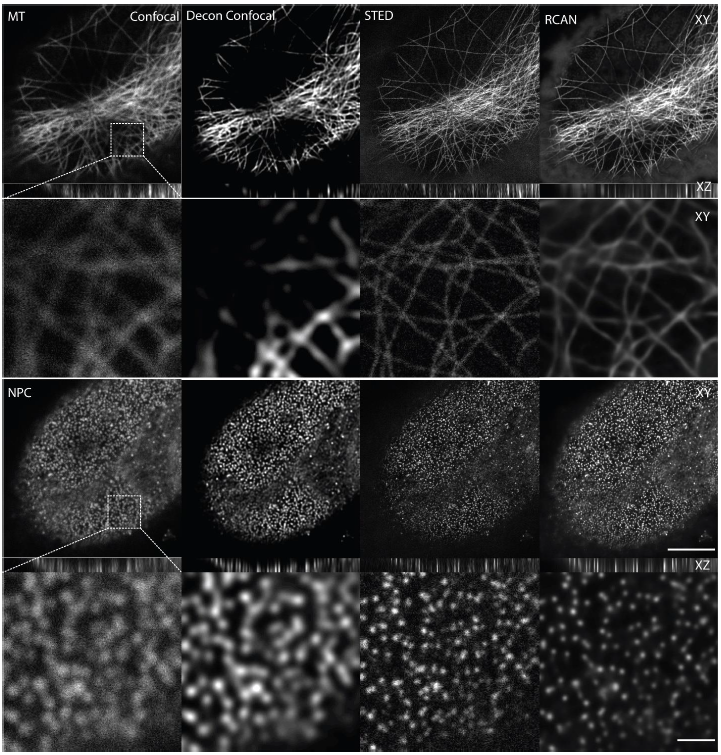
The trained #3DRCAN was applied to raw confocal data (incl. nuclear pores, microtubules and DNA), greatly improving (~2-fold) the spatial resolution. Imaging done with a Leica SP8. @LeicaMicro
#STED #superresolution #aimicroscopy #microscopy
9/17
#STED #superresolution #aimicroscopy #microscopy
9/17

Ultimately, we wanted to perform fast 4D confocal imaging and "convert" the raw data to STED-like images. So, we applied the trained #3DRCAN to live cell 3D (confocal) data sets. See 👇
#STED #superresolution #aimicroscopy #microscopy
10/17
#STED #superresolution #aimicroscopy #microscopy
10/17
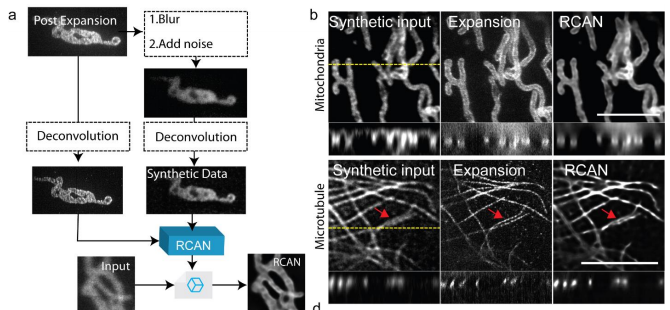
Lastly, we tested whether it was possible to further increase the resolution of iSIM (itself a #superresolution microscopy modality). To do this, we used expansion microscopy to generate the GT we needed to train #3DRCAN for this task.
#iSIM #ExM #aimicroscopy #microscopy 11/17
#iSIM #ExM #aimicroscopy #microscopy 11/17
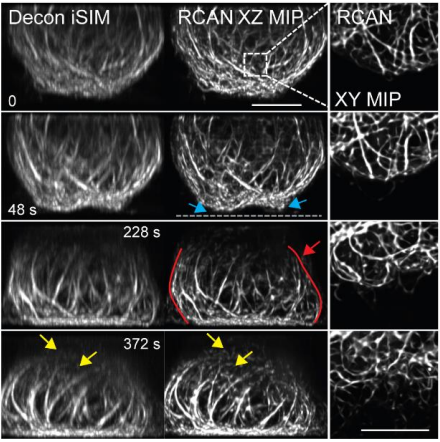
#3DRCAN was shown to improve both the lateral and axial resolution of iSIM images. Moreover, the improvement in resolution was superior to deconvolving the raw images.
#iSIM #ExM #superresolution #aimicroscopy #Microscopy 12/17
#iSIM #ExM #superresolution #aimicroscopy #Microscopy 12/17

As with the other key experiments, we successfully tested out the trained #3DRCAN on live cell 3D data sets. In this case we imaged mitochondria and microtubules using iSIM.
#iSIM #ExM #superresolution #aimicroscopy #Microscopy 13/17

#iSIM #ExM #superresolution #aimicroscopy #Microscopy 13/17

See all the supplementary information and videos here: biorxiv.org/content/10.110…
#microscopy #aimicroscopy #aivia #STED #iSIM #ExM #bioimageanalysis #deeplearning #3DRCAN
14/17

#microscopy #aimicroscopy #aivia #STED #iSIM #ExM #bioimageanalysis #deeplearning #3DRCAN
14/17


Disclaimer: this being a twitter thread, a lot of the details are missing - you are most welcome to review the pre-print. Your suggestions and input are welcome.
15/17
15/17
You can train your own #3DRCAN models and/or test out the ones we have described here. See github.com/AiviaCommunity… There are also some test images there. Let us know if you have any questions and/or suggestions.
#python #opensource #openaccess #OpenScience #aimicroscopy
16/17
#python #opensource #openaccess #OpenScience #aimicroscopy
16/17
Note to Aivia users: in the very short term all the models described in the #3DRCAN preprint will be available in #Aivia, #AiviaWeb and #AiviaCloud.
17/17
17/17
• • •
Missing some Tweet in this thread? You can try to
force a refresh