We are not far away from routinely #genome sequencing every newborn suspected of having a rare/hereditary disorder.
In fact, there will be a point, especially in single-payer health care systems, where it'll be logistically preferable to routinely #genome sequence every newborn.
In fact, there will be a point, especially in single-payer health care systems, where it'll be logistically preferable to routinely #genome sequence every newborn.
https://twitter.com/sbarnettARK/status/1338983774269415425
What will it take? Management, machines, and money (3M), in that order:
1/ A management system that handles sample collection, carries on the sequencing effectively, and makes the result available: we are not far away from this in, e.g. the UK's (@NHS+Wales/Scotland/NI).
1/ A management system that handles sample collection, carries on the sequencing effectively, and makes the result available: we are not far away from this in, e.g. the UK's (@NHS+Wales/Scotland/NI).
We are much better at this now than, say, 1-2 years ago. See a successful system of coordinating this for lower throughput sequencing but high numbers of samples in #COVID19 cogconsortium.uk
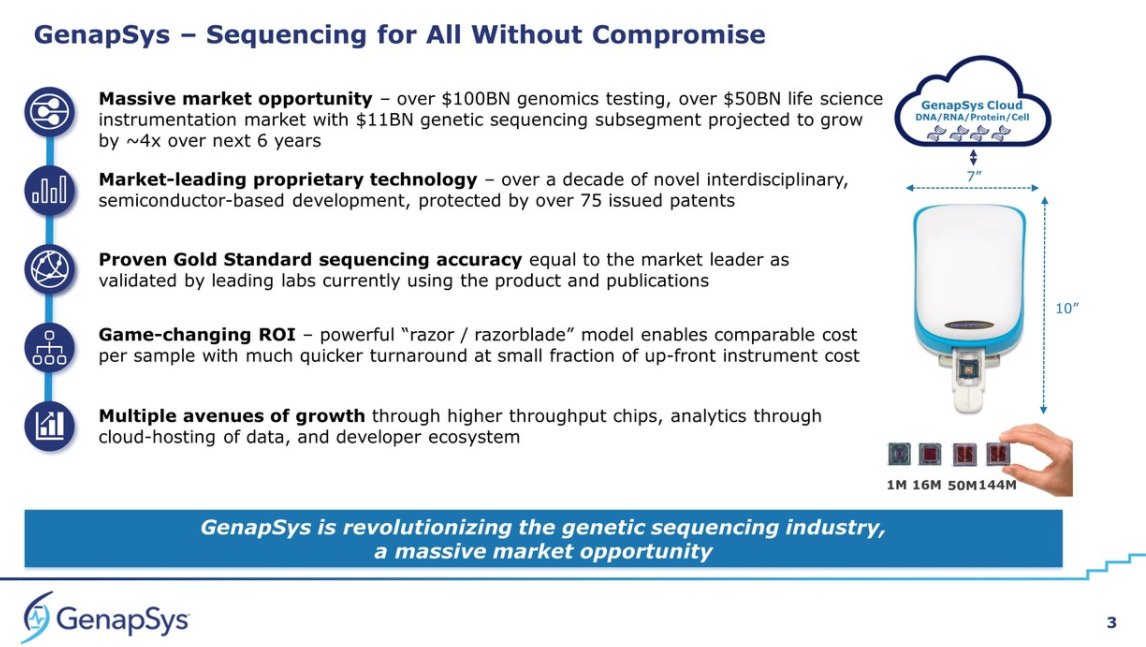
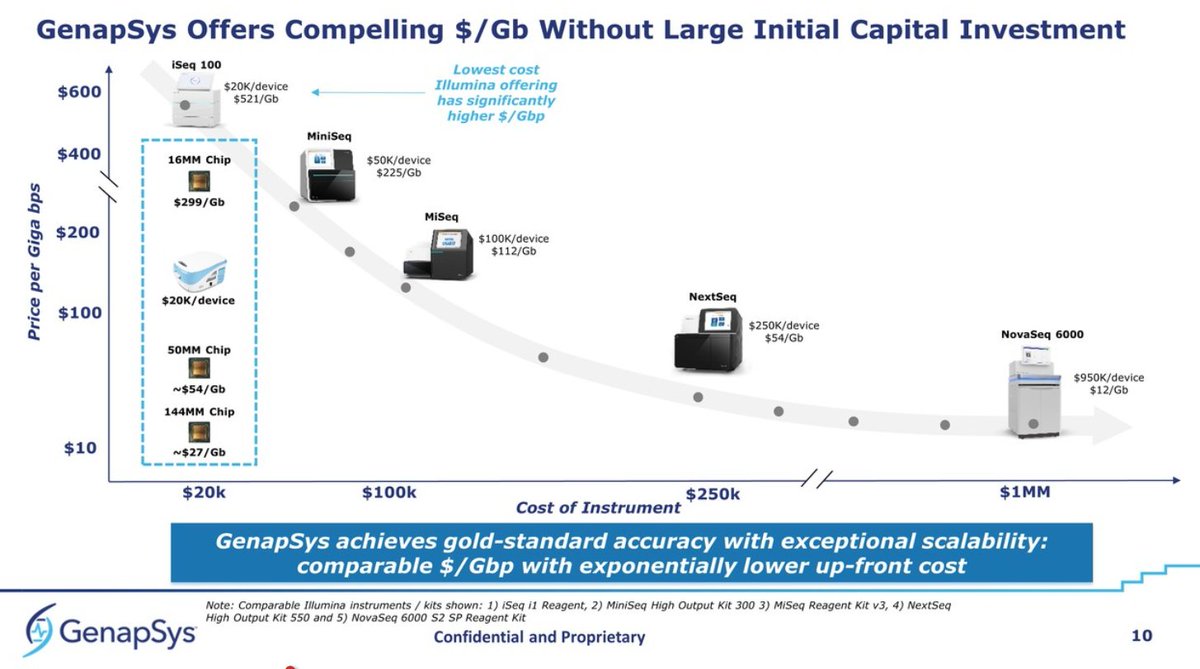
2/ Machines: we need higher global throughput of #NGS than currently: bit.ly/ngsspecs. Even if dedicating current install base #NovaSeqs to newborn sequencing, we would still need 8.2x more machines to keep up worldwide. But @MGI_BGI, @PacBio and @nanopore will grow up. 

E.g.
A/ @MGI_BGI has shown new tech that's many-fold higher throughput than a #NovaSeq (#DNBSEQTx / Surface coating tech).
B/ @nanopore #PromethION48 can now produce 1.0-1.2x the throughput of a #NovaSeq and the machine/flowcell production can be scaled up significantly.

A/ @MGI_BGI has shown new tech that's many-fold higher throughput than a #NovaSeq (#DNBSEQTx / Surface coating tech).
B/ @nanopore #PromethION48 can now produce 1.0-1.2x the throughput of a #NovaSeq and the machine/flowcell production can be scaled up significantly.


C/ @Pacbio #Sequel2e and iterations on path to produce lots of contiguous, haplotype-aware genome assemblies (see results from @lh3lh3 Hifiasm et al.)
D/ Other technologies can be used to "contiguate" / fill-in the gaps from short-reads, e.g. @bionanogenomics (see @genomeark).
D/ Other technologies can be used to "contiguate" / fill-in the gaps from short-reads, e.g. @bionanogenomics (see @genomeark).
So a possible scenario: a single-payer system (e.g. @NGS or #MedicareForAll) where the newborn's genome is sequenced to high quality, and results are available to the health system during their entire lives (from cradle to grave omics). From then onwards, the info is used
to check for newborn/infant rare diseases (0-1yo), children's cancer (1-5yo), testicular cancer (15-30yo), colorectal cancer screening (45+yo), etc. Lots of benefits to the system for regular #LiquidBiopsy monitoring using #epigenomic profiling.
Then there is a second wave of Precision Medicine and Omics that will benefit from large-scale Population Genomics. I have to confess that I lost touch with these in the last few years, so someone else will have to give colour to these here.
Again, this is not far away, and as George Church described in a recent podcast: we are now underestimating scientific advances in the general public, compared to the perception we had in the 1950-60s, so all this is happening, and will be part of our daily lives soon.
@d2unroll unroll
*@NHS or #MedicareForAll, or equivalents in every geography.
• • •
Missing some Tweet in this thread? You can try to
force a refresh