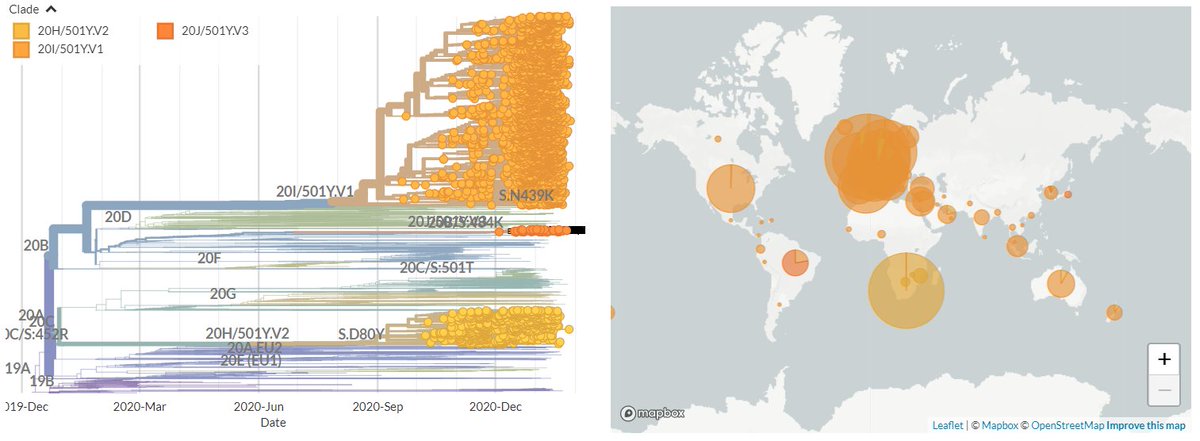
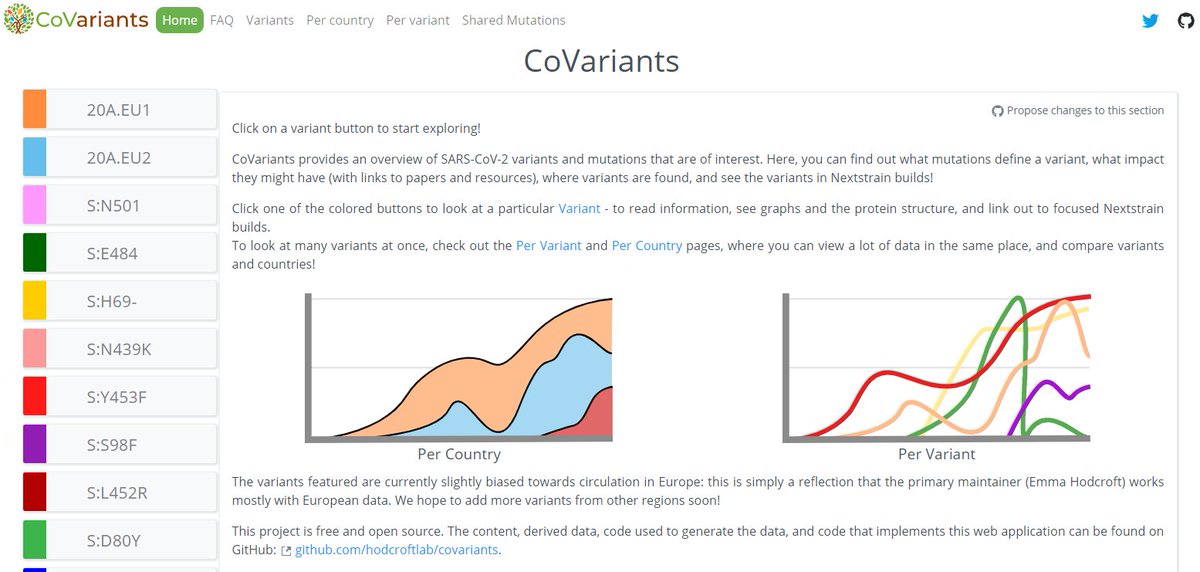
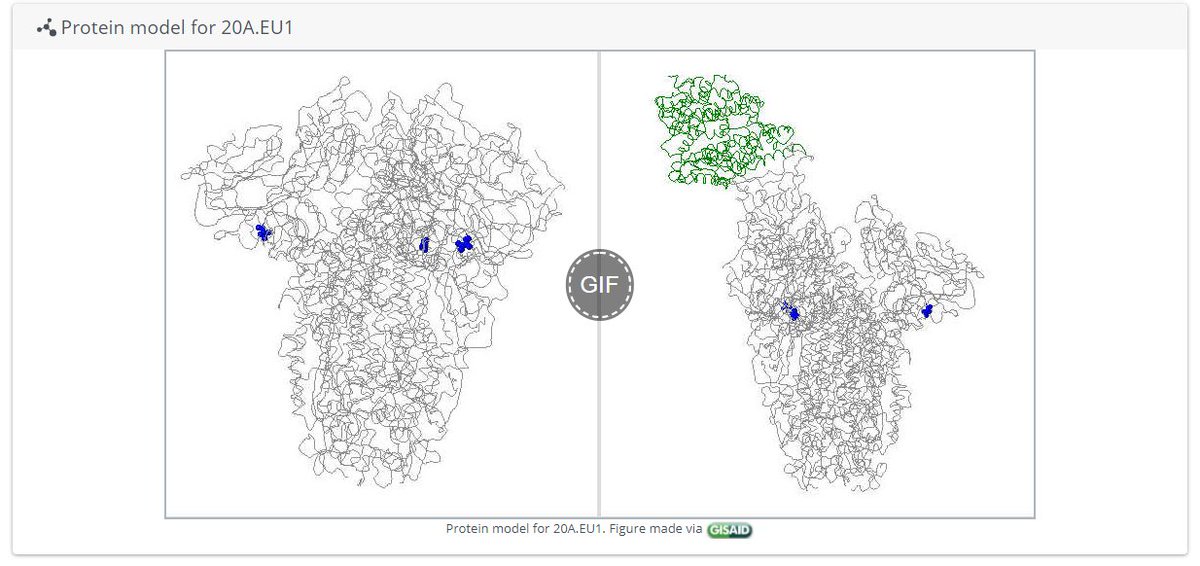
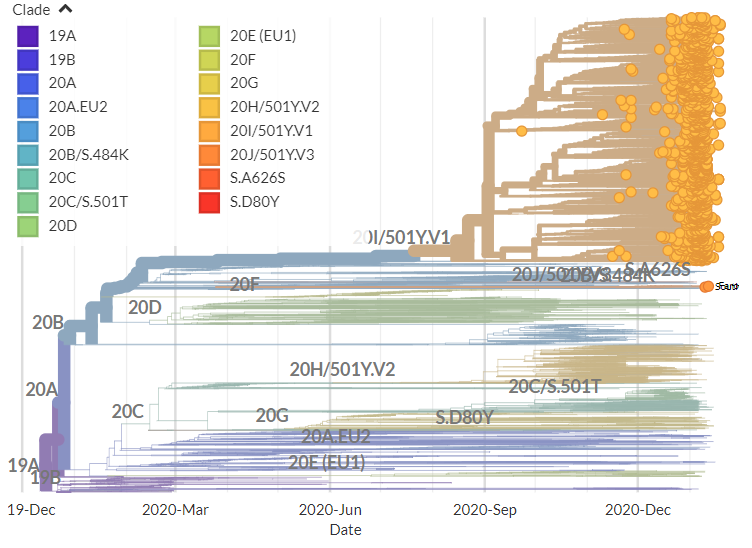
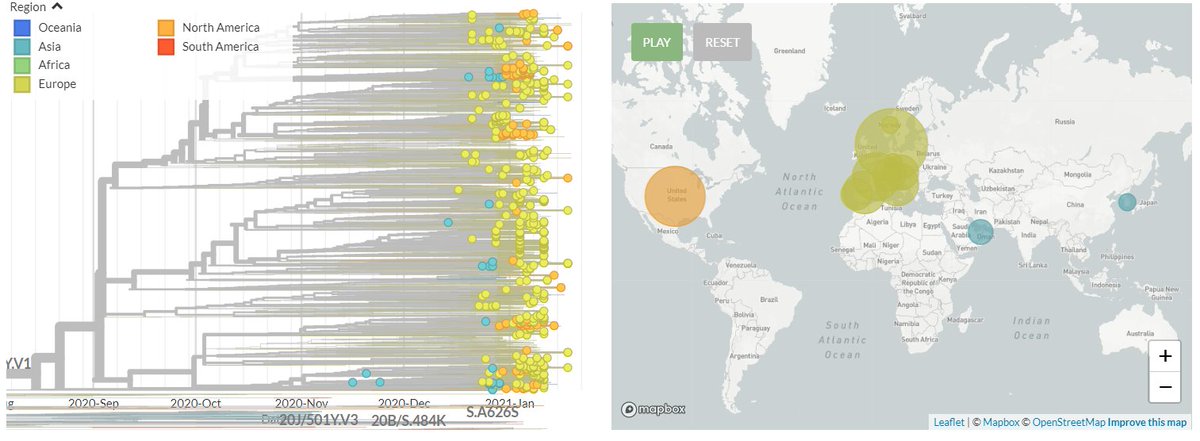
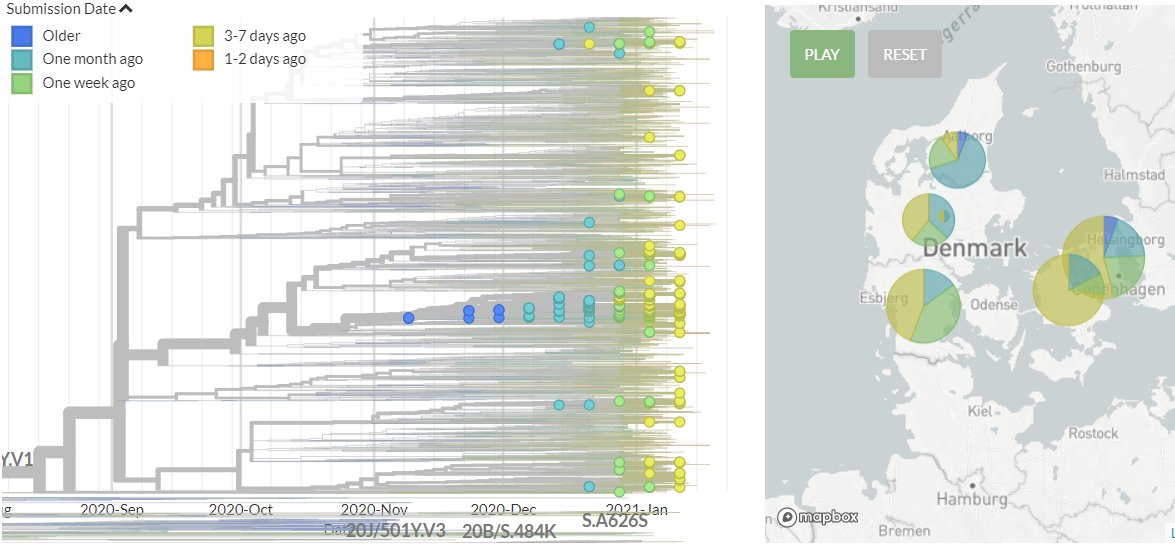
The focal builds for S:N501 & S:E484 are now updated to data from the 8th Feb!
nextstrain.org/groups/neherla…
nextstrain.org/groups/neherla…
Let's take a quick look at some snapshots of what we can see in the latest data.
1/15
nextstrain.org/groups/neherla…
nextstrain.org/groups/neherla…
Let's take a quick look at some snapshots of what we can see in the latest data.
1/15

Important note: the S:N501 build is now being more significantly impacted by downsampling.
I'm working on addressing this, but in the meantime, suggest looking at the S:E484 build for 501Y.V2 & 501Y.V3 & checking out the CoV Lineages report! cov-lineages.org/global_report.…
2/15
I'm working on addressing this, but in the meantime, suggest looking at the S:E484 build for 501Y.V2 & 501Y.V3 & checking out the CoV Lineages report! cov-lineages.org/global_report.…
2/15
Israel now has over 200 sequences in 501Y.V1 (B.1.1.7). The latest additions indicate separate introductions (mostly from a month or two ago) and a large and still-growing local cluster.
3/15

3/15


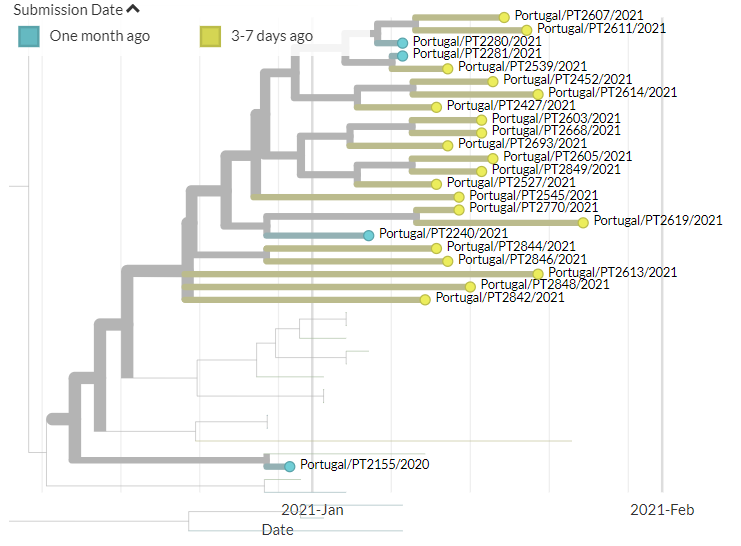
Portugal has over 170 sequences in 501Y.V1 (B.1.1.7) now. Many are separate introductions, but there are clusters that seem to indicate local transmission.
4/15

4/15

Sweden now has over 60 samples in 501Y.V1 (B.1.1.7). These indicate both separate introductions and signs of local transmission in clusters.
5/15

5/15


Turkey now has over 100 sequences in 501Y.V1 (B.1.1.7), indicating both local transmission and multiple separate introductions.
6/15

6/15


France now has over 44 samples in 501Y.V2 (variant predominant in South Africa), including continued growth of a large, locally transmitted cluster.
7/15

7/15


Belgium has over 30 samples in 501Y.V2 (variant predominant in South Africa) now, mostly indicating separate introductions, but some small clusters, that could indicate limited local transmission or exposure to a common source.
8/15

8/15


Botswana now has 15 sequences in 501Y.V2 (variant predominant in South Africa), indicating multiple introductions, and either exposure to a common source or local transmission.
9/15

9/15


Belgium now has almost 40 sequences in 501Y.V2 (variant predominant in South Africa). Most of these seem to fall into a large cluster that indicates ongoing local transmission.
10/15

10/15


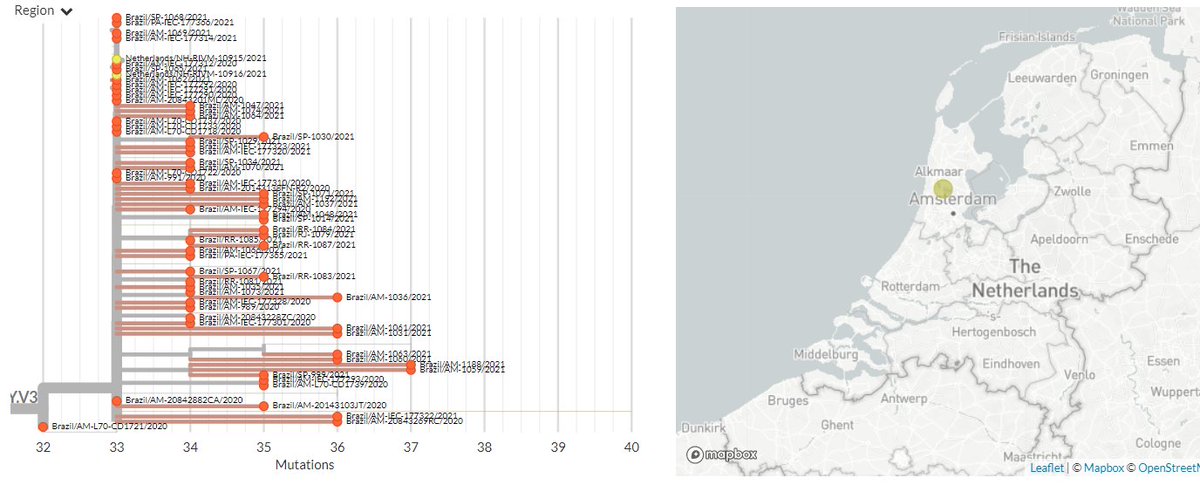
There are 2 sequences from the Netherlands in 501Y.V3 (P.1) now. These are identical, so could be from a common exposure.
11/15
11/15
France has 3 sequences in 501Y.V3 (P.1). Remember, vertical position on a horizontal line is not meaningful, so these could be from 1 exposure or introduction.
12/15
12/15

The USA also has 3 sequences in 501Y.V3 (P.1), two in Minnesota which seem to be linked, and one from Oklahoma, a separate introduction.
13/15
13/15

The USA also has almost 40 sequences in 20B/S:484K (not currently a variant of concern) now, showing multiple introductions and some clusters that may indicate ongoing transmission.
14/15
14/15

The Netherlands now has 6 sequences in 20B/S:484K (not currently a variant of concern), indicating additional independent introductions.
15/15
15/15

There's a mistake earlier in the thread! Corrected:
*The Netherlands* has over 30 samples in 501Y.V2 now, mostly indicating separate introductions, but some small clusters, that could indicate limited local transmission or exposure to a common source.
16/15

*The Netherlands* has over 30 samples in 501Y.V2 now, mostly indicating separate introductions, but some small clusters, that could indicate limited local transmission or exposure to a common source.
16/15


• • •
Missing some Tweet in this thread? You can try to
force a refresh