
1/n Weekly #SARSCoV2 variant report from Connecticut - data from @YaleSPH, @jacksonlab, and @CTDPH.
- B.1.1.7 = 63 cases
- B.1.351 = 1 case
- P.1 = 0 cases
B.1.1.7 cases by county 👇. Most in New Haven due to surveillance biases.
Report:
covidtrackerct.com/variant-survei…
- B.1.1.7 = 63 cases
- B.1.351 = 1 case
- P.1 = 0 cases
B.1.1.7 cases by county 👇. Most in New Haven due to surveillance biases.
Report:
covidtrackerct.com/variant-survei…

2/n
Temporal distribution of B.1.1.7 cases doesn't accurately reflect the growth of B.1.1.7 in CT as the sequence reporting is still lagging. Next week we'll have temporal est of frequency based on SGTF. B.1.1.7 is currently associated with 5-10% of the COVID-19 cases in CT.
Temporal distribution of B.1.1.7 cases doesn't accurately reflect the growth of B.1.1.7 in CT as the sequence reporting is still lagging. Next week we'll have temporal est of frequency based on SGTF. B.1.1.7 is currently associated with 5-10% of the COVID-19 cases in CT.

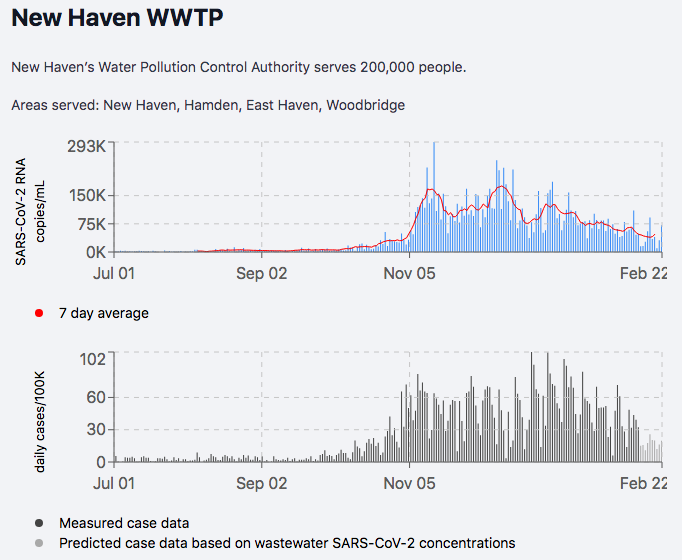
4/n While B.1.1.7 transmission is firmly established in CT, overall transmission of SARSCoV2 continues to decline. Shown by wastewater and cases (New Haven). As B.1.1.7 becomes more dominant, it *may* reverse this trend.
yalecovidwastewater.com/reports
yalecovidwastewater.com/reports
5/n We've detected 7 cases of B.1.429 in CT (first detected in California). This is not yet an official variant of concern, but it is one of many that we are watching. Much more research is needed to back up some of the claims about B.1.429. 

6/n We've detected 2 lineages in CT with the E484K mutation - B.1.525 (8 cases) and the now infamous B.1.526 "New York variant" (11 cases). The concern is that E484K *may* decrease vaccine efficacy, but we really don't know what if does on these lineages. MUCH more data needed. 

7/7 As always - huge thanks to our team for setting up a system to help conduct genome surveillance for our state! @JosephFauver, @tdalpert, @MaryPetrone10, @VogelsChantal, @mallerybreban, @aewatkins6, @IsabelOtt
The analysis and reports by:
@ChaneyKalinich & @AndersonBrito_
The analysis and reports by:
@ChaneyKalinich & @AndersonBrito_
• • •
Missing some Tweet in this thread? You can try to
force a refresh









