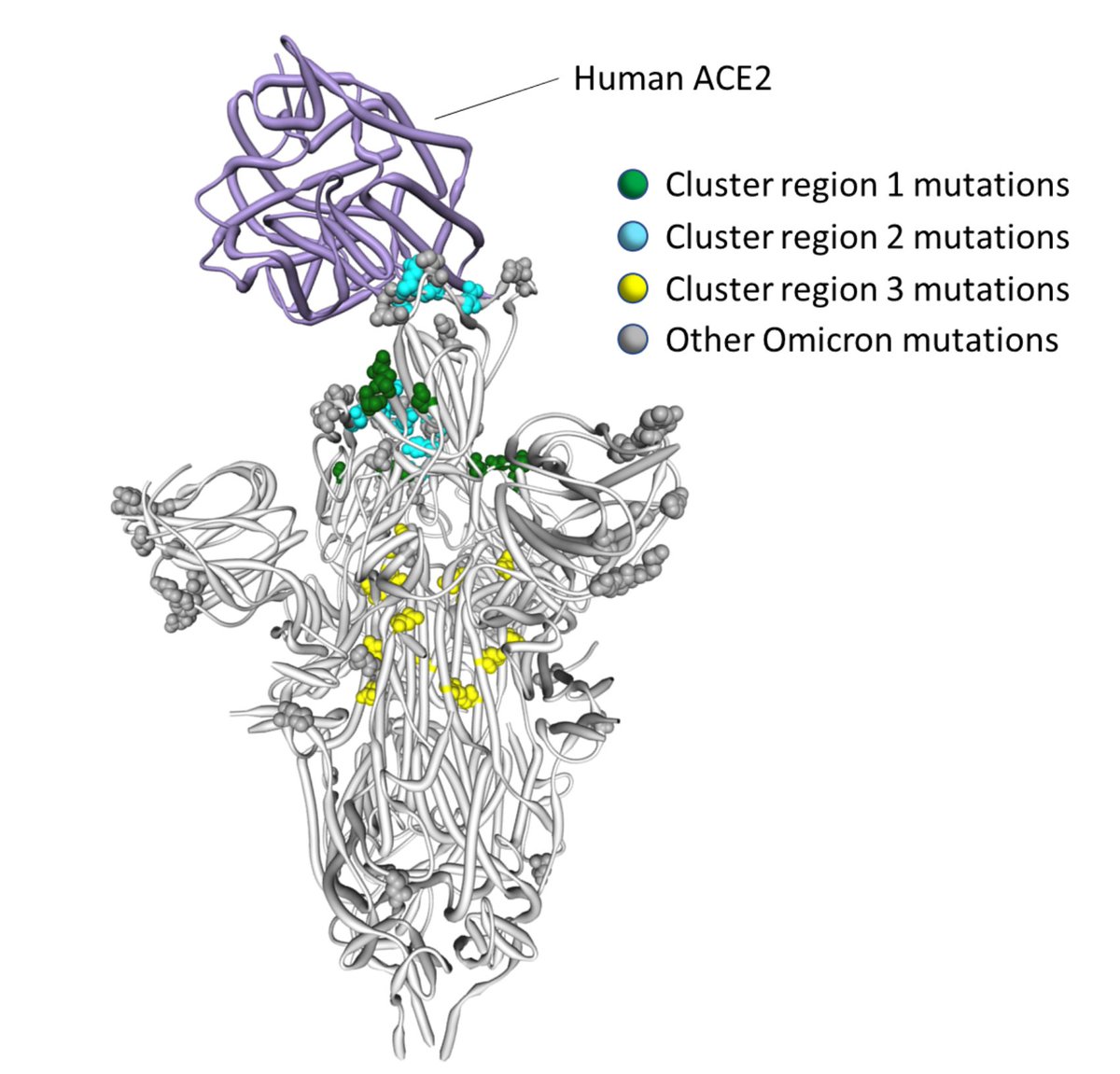
1/8 Selection analysis update on the S-gene of #Omicron. Caution: preliminary data and analysis; based on ~800 sequences (12/7) from @GISAID classed as B.1.1.529 by #Pangolin. There appears to be signal of positive selection in circulating #Omicron sequences. @DarrenM98230782
2/8 Thanks to all the data contributors, including @Tuliodna and SA colleagues! We find evidence of positive selection at S/346 (K), S/701 (V), S/211 (several states, possibly due to indel alignment issues). Genomes have a lot of noise around S/214, so this could be an artifact.
3/8 Weaker signals are also present at some of the sites that have been selected during the emergence of #Omicron (S/371,S/375, S/505), and sites in the RBD (S/440, S/446,S/452).
4/8 Mutations at S/346 have been found in other lineages (e.g. mu), and are known to reduce antibody binding for certain MAbs (covdb.stanford.edu/search-drdb/?h…)
5/8 A mutation at S/701 is a part of the N501Y metasignature that we developed for alpha, beta and gamma lineages (cell.com/cell/pdf/S0092…). These are sites where lineages carrying N501Y mutations have previously shown evidence for convergent evolution => towards similar targets
6/8 #Omicron might be exploring some of the same evolutionary routes as those taken by previous N501Y carrying lineages. Site S/716 (also a part of our meta-signature) is showing marginal evidence for selection.
7/8 There is apparent ongoing evolution at the sites that are newly mutated in #omicron (371,375,505). We hypothesized that these sites might be involved in epistatic interactions that developed during unobserved evolution of this lineage (virological.org/t/selection-an…)
8/8 Continuing changes at these sites might indicate that further adaptation/reversion is place as the virus is spreading globally. As evolution progresses, we will be able to better resolve which it is.
• • •
Missing some Tweet in this thread? You can try to
force a refresh