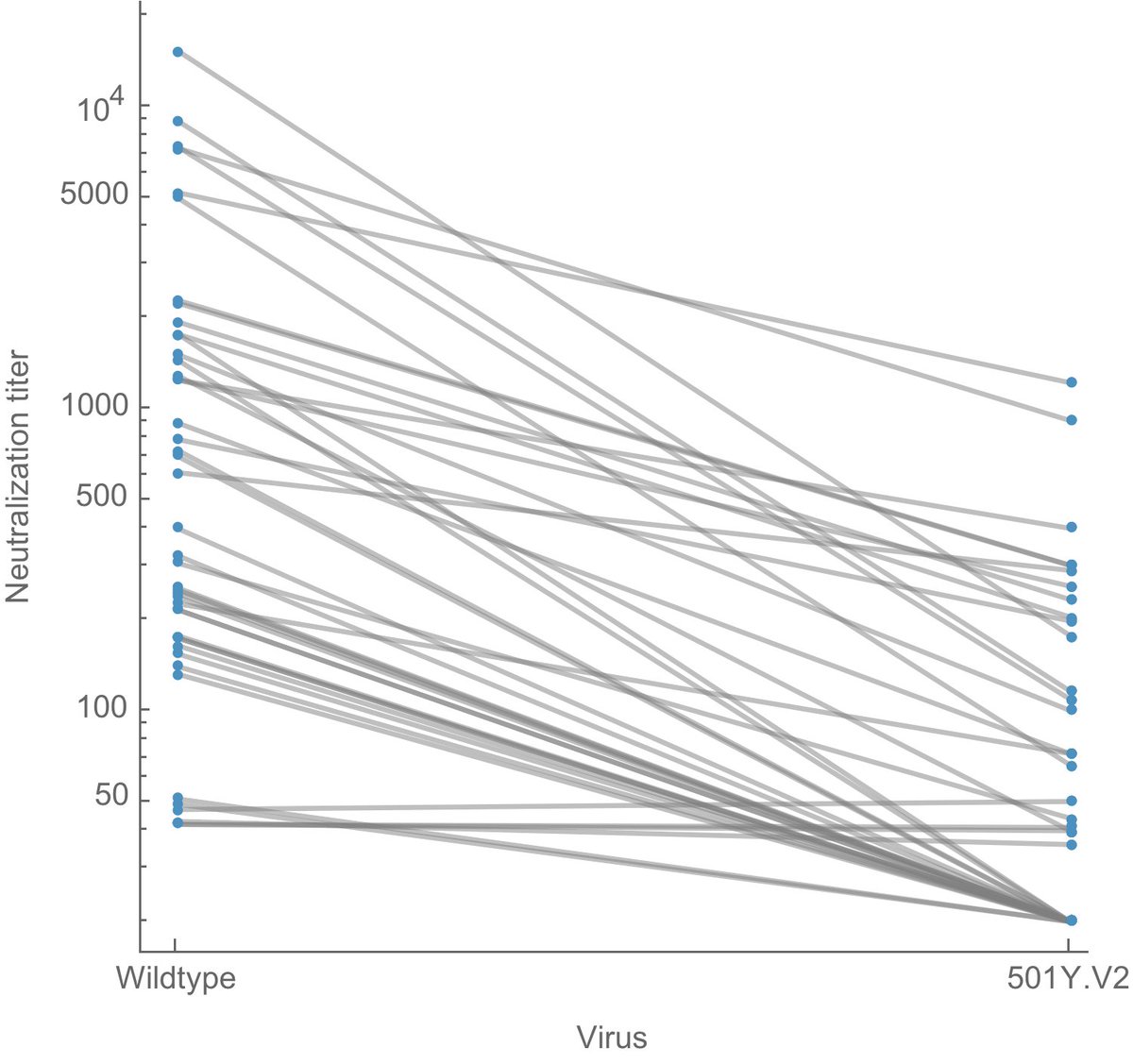
There are effectively two #COVID19 epidemics in the US at this moment; one largely resolving epidemic comprised of non-variant viruses and one growing epidemic of B.1.1.7. Together they have resulted in a near-plateau of cases throughout much of the spring. 1/10
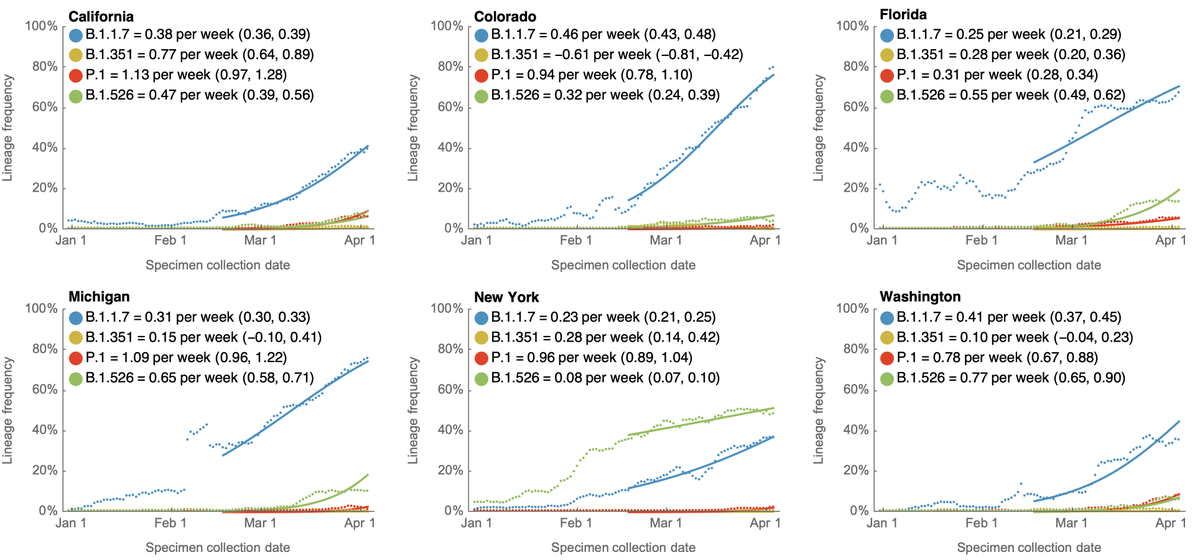
If we look at virus frequencies in the US using data in @GISAID, we can see that the 7-day weighted frequency of B.1.1.7 has been growing consistently since January and is now over 50% in the US. 2/10 

This pattern is repeated across individual states. These six were chosen as states with plentiful genomic data and to provide geographic diversity. B.1.1.7 is dominating throughout the US, except for New York and surroundings where B.1.526 is co-circulating. 3/10 
We can use the estimated frequency of B.1.1.7 to split cases reported by @CDCGov (data.cdc.gov/Case-Surveilla…) into non-B.1.1.7 cases and B.1.1.7 cases. In the following I've applied a 7-day smoothing to case trajectories. 4/10
This is showing B.1.1.7 cases in blue stacked on top of non-B.1.1.7 cases in gray, where it's clear that non-B.1.1.7 cases have continued to decline, while B.1.1.7 cases have continued to increase. 5/10 
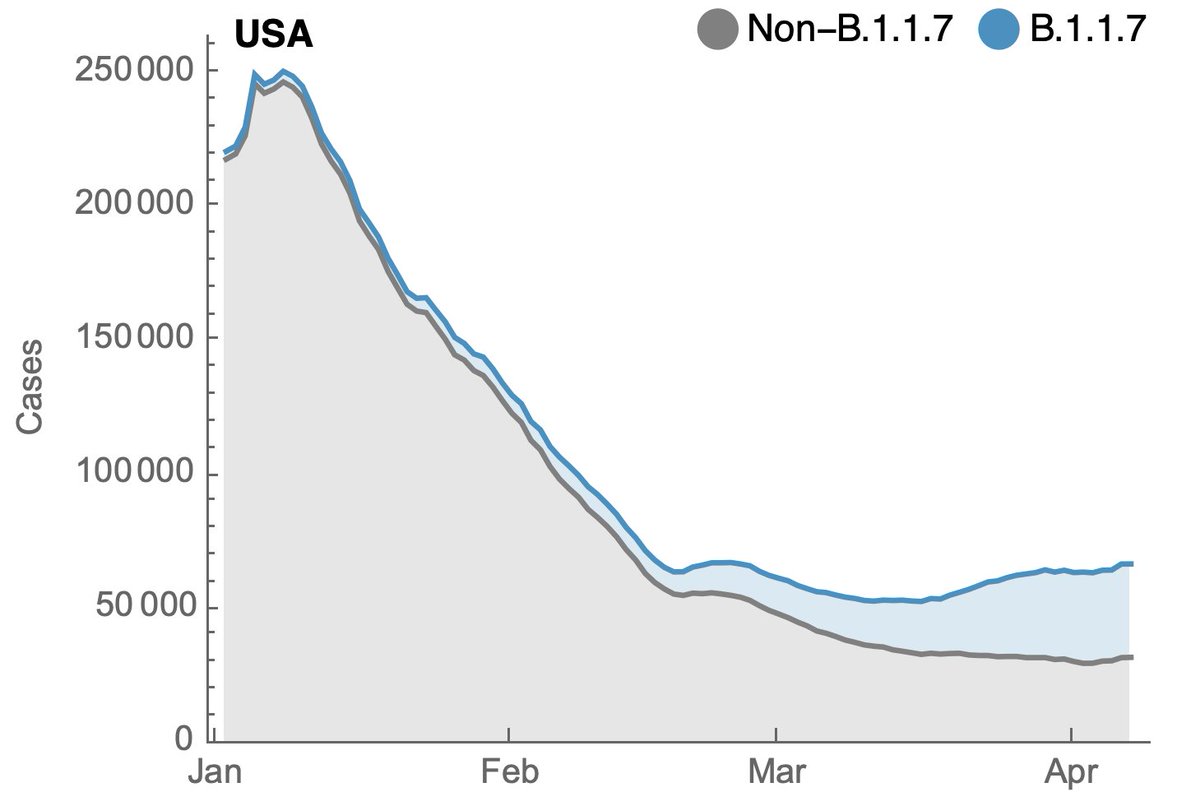
Plotting each on a log scale makes growth and decline more obvious. Without the contribution of B.1.1.7 there would have been ~32k cases on April 7 rather than the observed ~65k cases. 6/10 

We can see the same pattern repeated in individual states with growing B.1.1.7 epidemics on top of a largely resolving non-B.1.1.7 background. Some states like California have less of a B.1.1.7 epidemic and some like Colorado and Michigan have more of a B.1.1.7 epidemic. 7/10 

Again, plotting non-B.1.1.7 cases (in gray) and B.1.1.7 cases (in blue) on a log scale makes the separation of growth and decline more obvious. 8/10 
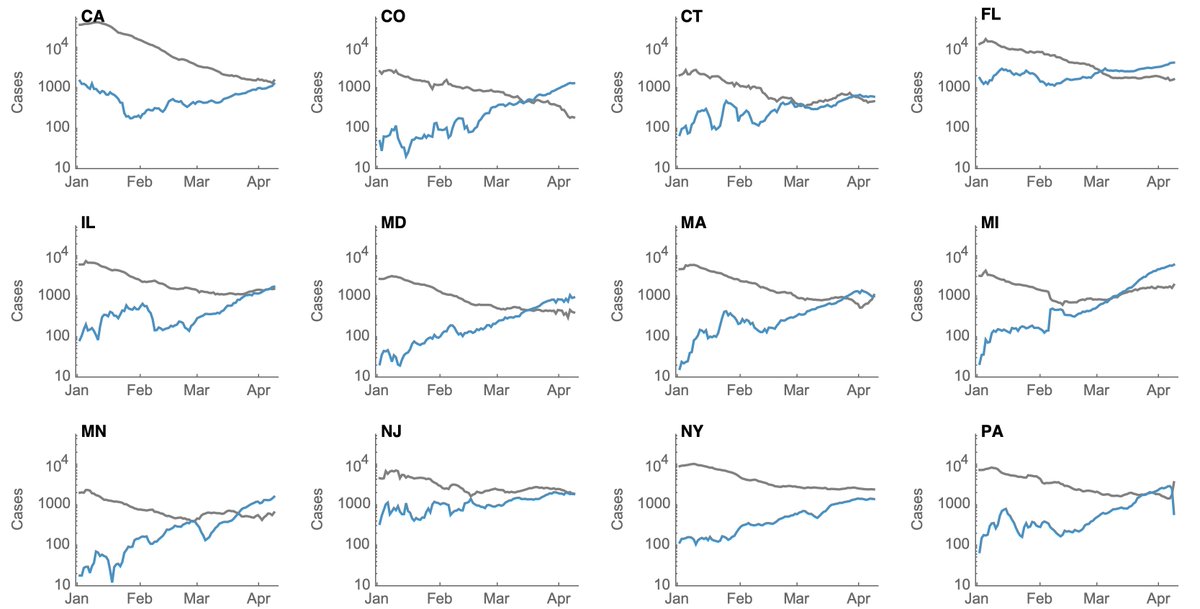
This is consistent with what's been seen in the UK and in Europe and supports a transmission advantage of B.1.1.7. Thus we see that the same conditions that can lead to control of older non-B.1.1.7 viruses and Rt < 1, can still result in growth of B.1.1.7 and Rt > 1. 9/10
That said, there are multiple factors that should be resulting in continued improvements to the US COVID-19 epidemic (vaccination, continued build up of natural immunity from infection, improved seasonality). It's unclear to me how much of a wave from B.1.1.7 to expect. 10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh