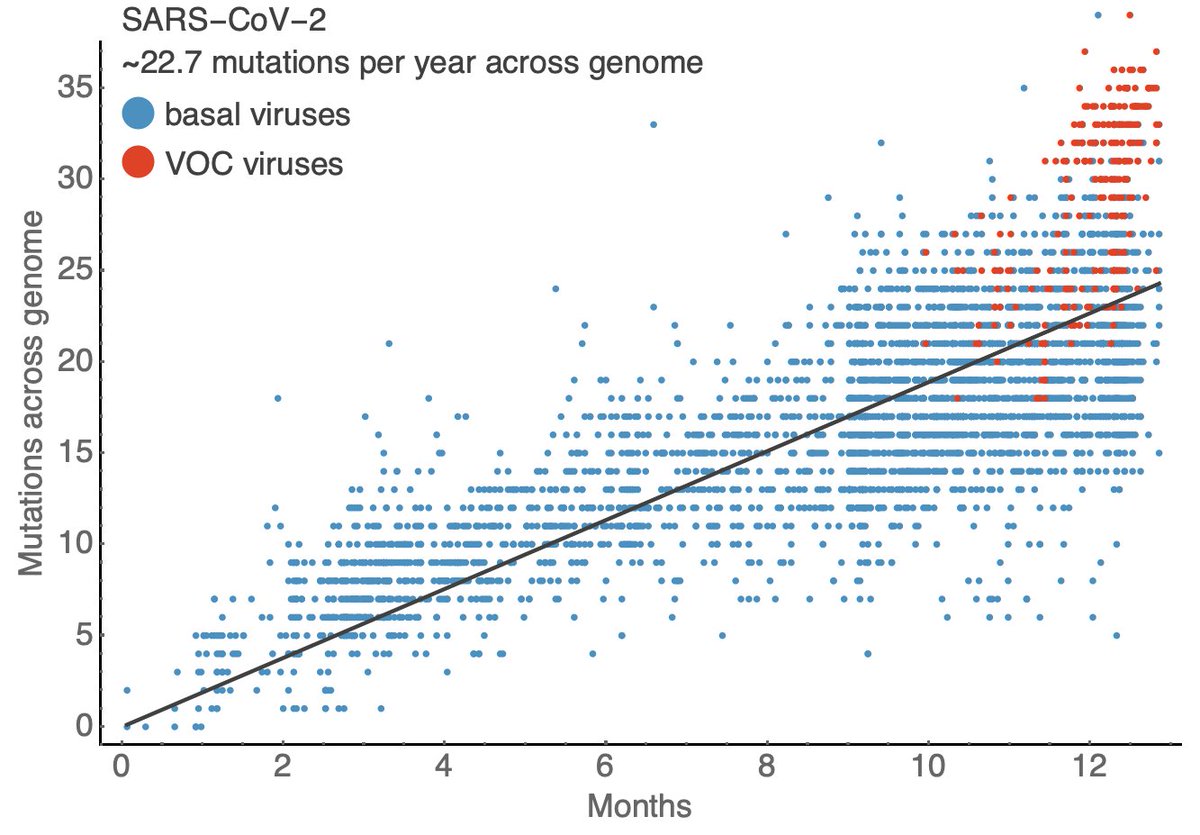
Just as we can decompose the US #COVID19 epidemic into a B.1.1.7 epidemic and a non-B.1.1.7 epidemic, we can further partition by variants of concern B.1.1.7, B.1.351 and P.1, where it's clear that P.1 has been gaining ground. 1/13
https://twitter.com/trvrb/status/1386694440023068677
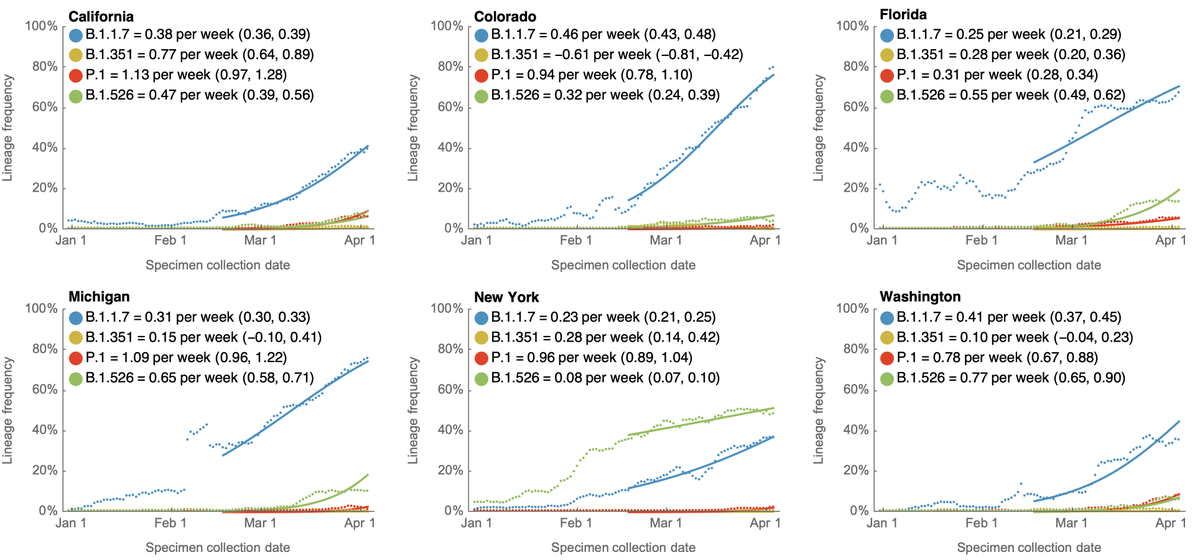
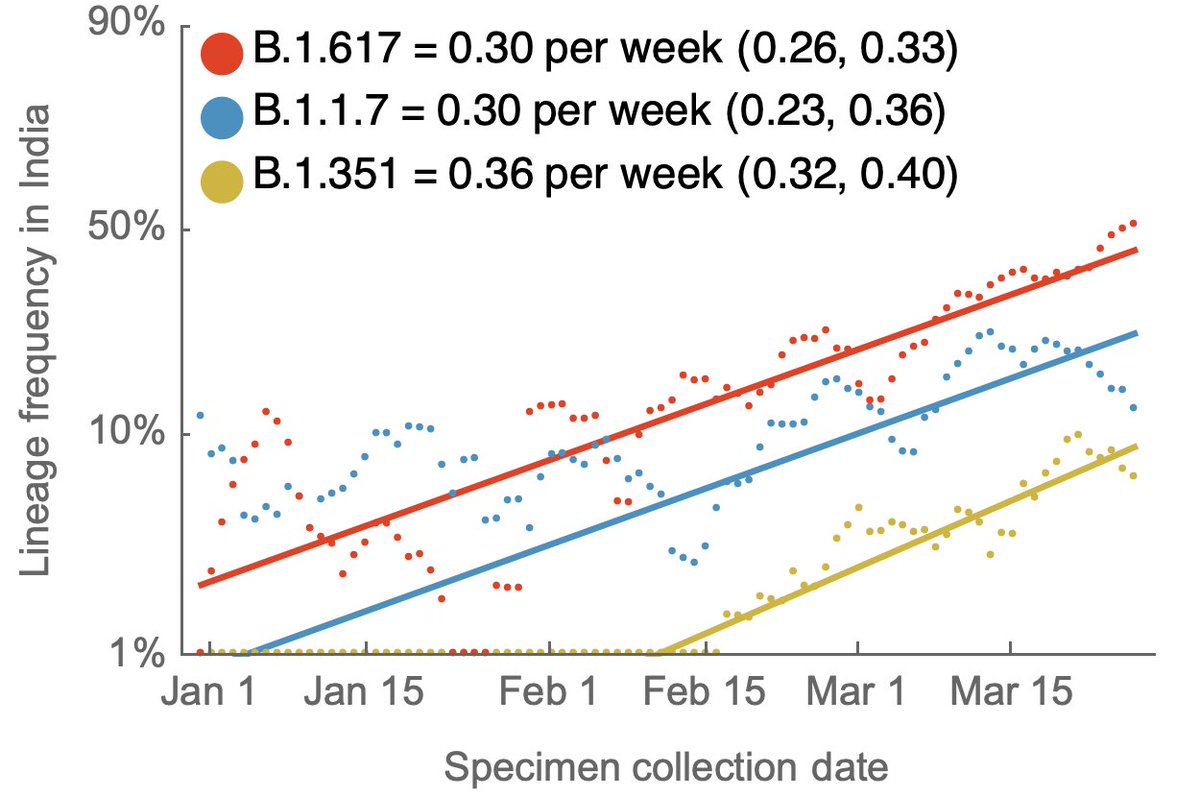
Here, using data from @GISAID, we see that in terms of frequencies across the US, P.1 has been undergoing more rapid logistic growth in frequency than B.1.1.7, while B.1.351 has been slower than B.1.1.7. 2/13 
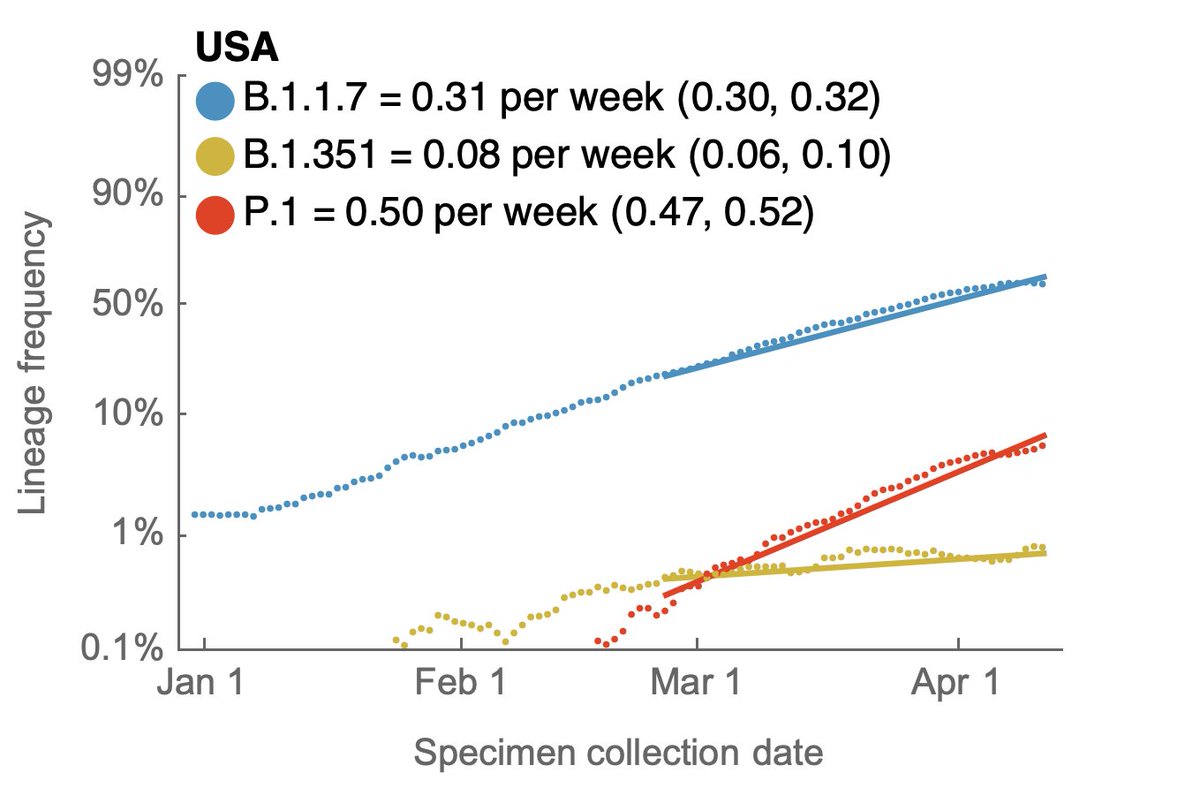
I'm plotting this with the unusual "logit" y-axis (with 1%, 10%, 50%, etc...) because a straight line in logit space is indicative of logistic growth. This sort of plot makes it easy to compare logistic growth rate of frequency between lineages with different frequencies. 3/13
We generally see more rapid logistic growth of P.1 across individual states as well. These 9 were chosen as those with the most data. All these plots only go to Apr 14, as there is a necessary lag to the genomic data as samples are processed. 4/13 

The observation of faster logistic growth of P.1 compared to B.1.1.7 suggests that P.1 may have a transmission advantage over B.1.1.7 in the US and may continue to gain ground even as B.1.1.7 comes to dominate the virus population. 5/13
Given that P.1 frequency is growing rapidly, but overall cases are generally falling, the question becomes are vaccines and other control measures enough to curb spread of P.1 in absolute case counts? 6/13
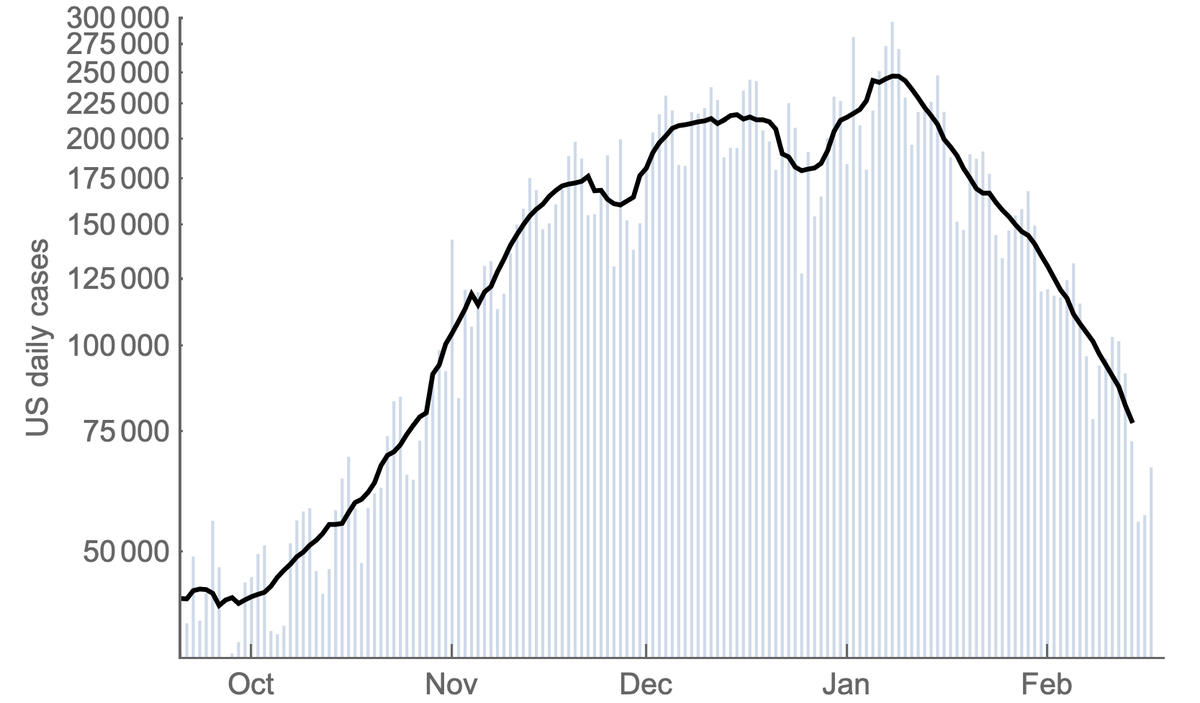
To address this we can take these frequency estimates alongside case counts from @CDCGov and use frequencies to partition case counts by variant. Doing so results in this plot for the US, where the large majority of the epidemic is due to B.1.1.7 and non-B.1.351/P.1 viruses. 7/13 
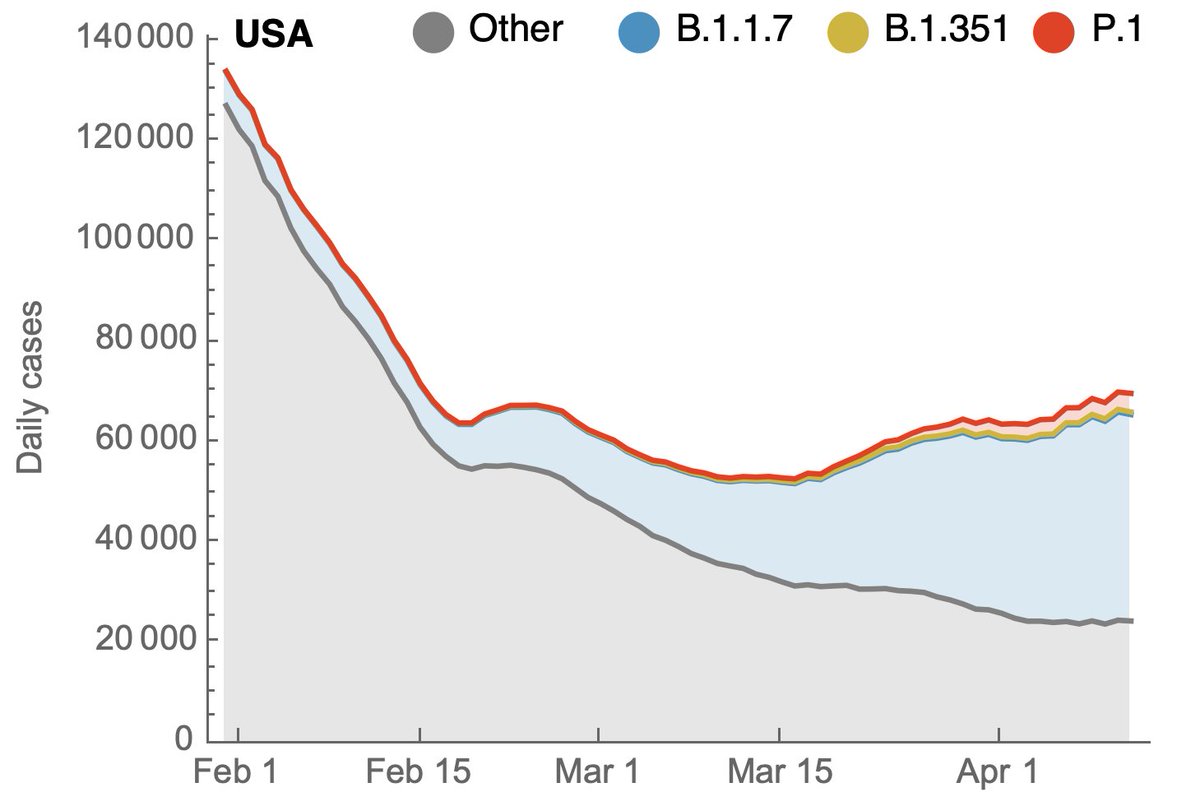
However, plotting partitioned case counts on a log-scale clearly shows the continued growth in absolute case counts of P.1. 8/13 

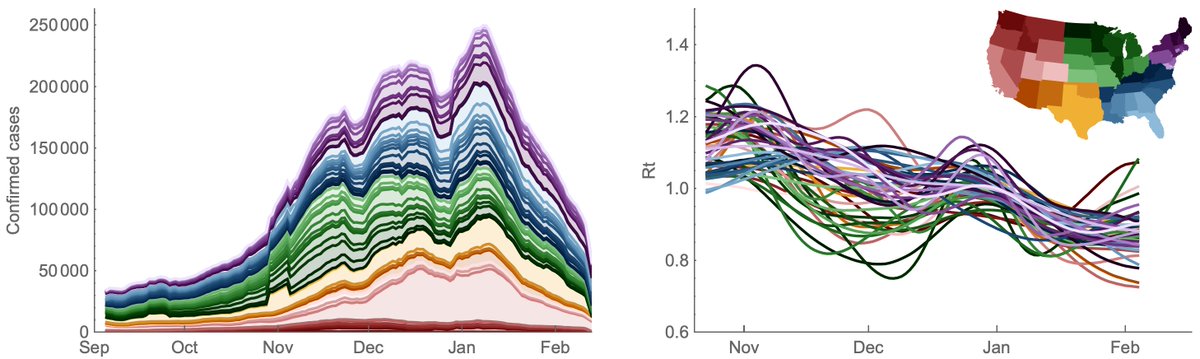
We can use the same approach with state-level case counts and frequency data to estimate case counts for B.1.1.7, B.1.351 and P.1 variants. Here, we see that P.1 is still a minor (but growing) contribution but where some states like IL and WA have a larger P.1 share. 9/13 

Looking at this same data on a log-scale makes the absolute growth of P.1 cases in CA, FL, IL, MA, MI, NY and WA quite clear. 10/13 

Vaccination will continue bring down overall transmission rate, where I expect transmission chains of non-variant viruses to largely die out in the coming weeks. Variant viruses B.1.1.7 and P.1 raise the bar for the level of vaccination required to control the epidemic. 11/13
Mutations in P.1 suggest partial escape from antibody binding, which is borne out in drops in neutralization titer. I would expect P.1 to show some decrease in vaccine effectiveness, but remain largely effective. Figure from Wu et al (nejm.org/doi/full/10.10…). 12/13 

Growth of P.1 alongside probable decrease in vaccine effectiveness suggests it's all the more important to get as many people vaccinated as possible as vaccination can still suppress circulation of P.1; it's just that the bar for herd immunity is higher. 13/13
• • •
Missing some Tweet in this thread? You can try to
force a refresh