#COVID19 cases in the US reported by @CDCGov have continued their week-after-week exponential decline that began in mid-April. This is exceptionally welcome news, although I'm now watching closely for variants driving sub-epidemics despite overall cases falling. 1/10 

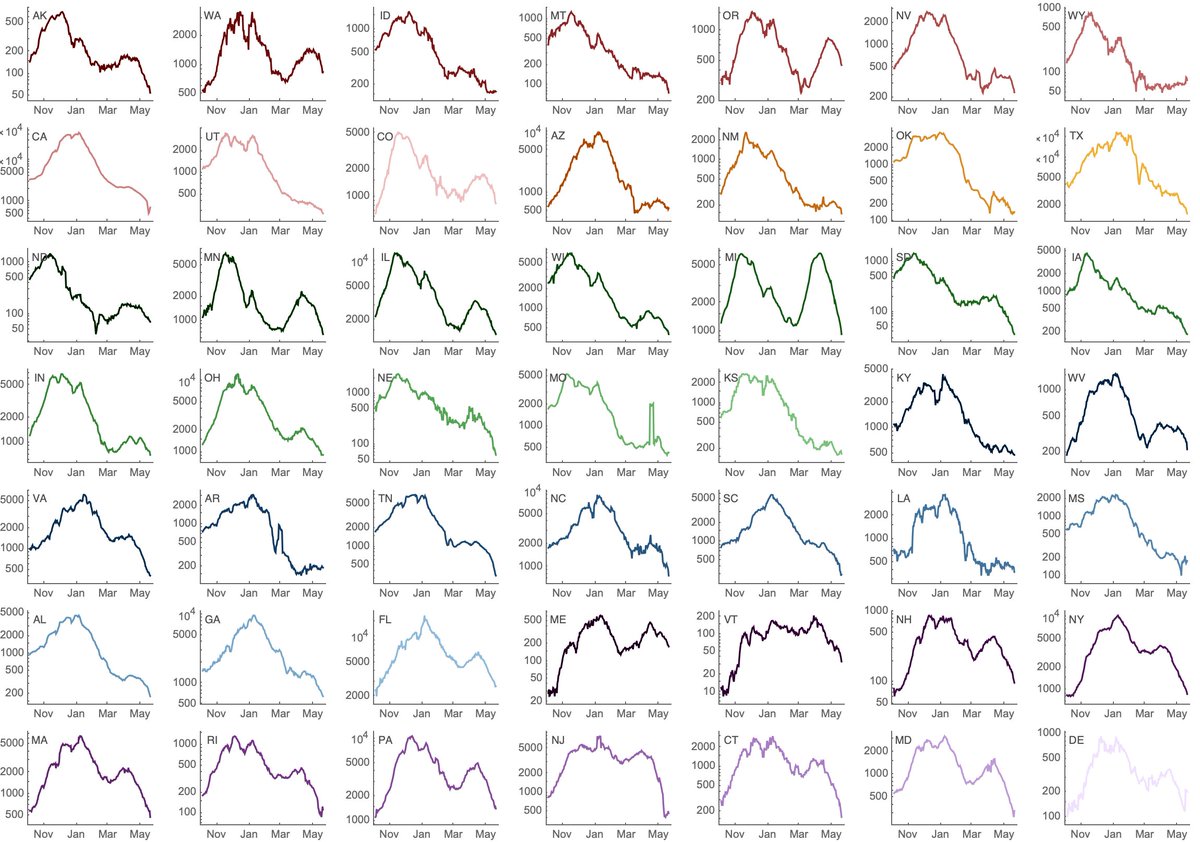
If we look at state-level cases with a log-axis we can see exponential growth and then exponential decline visible as straight lines on the log plot. Some states have had recent precipitous declines (NY, MA, MI), while others have been more stable (WA, CO, OR). 2/10 
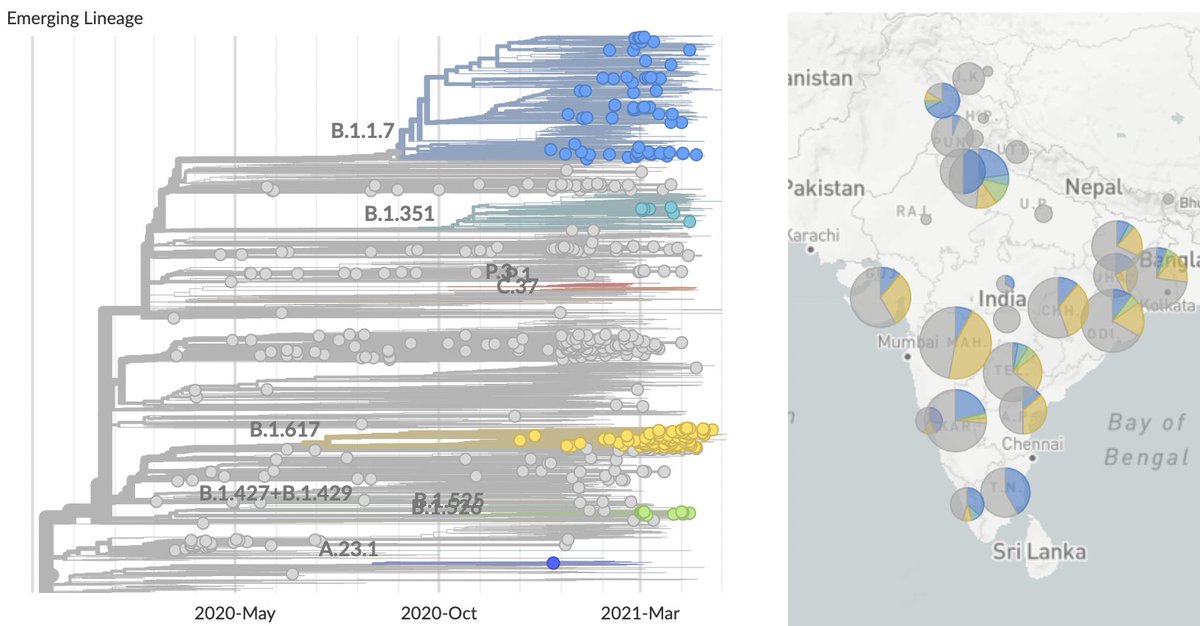
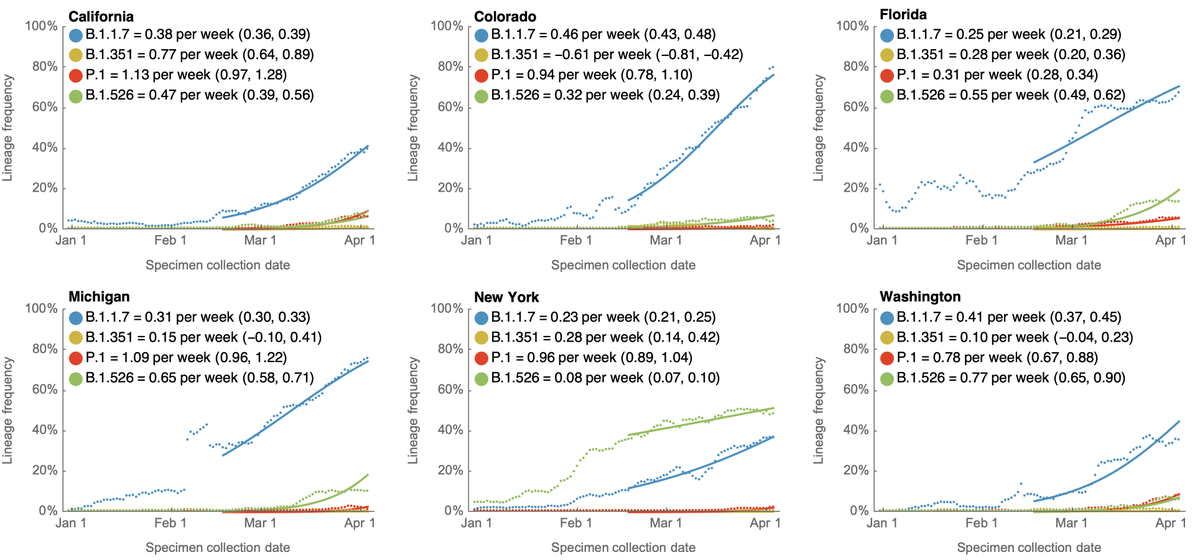
Using genomic data shared to @GISAID, we can plot frequency of different variant lineages through time and across states to get a sense of competitive dynamics. Here, I'm plotting lineage frequency on a logit axis, so that logistic growth is visible as a straight-line fit. 3/10 

Relative fitness becomes more clear as variants are placed in direct competition. B.1.1.7 and B.1.526 had been increasing rapidly in frequency in New York. However, since they reached high frequency they've been in more direct competition and B.1.1.7 has edged out B.1.526. 4/10 

I'm now watching Illinois in particular where P.1 is at ~30% frequency and B.1.1.7 is at ~60% frequency to see if P.1 starts to displace B.1.1.7 there. 5/10 

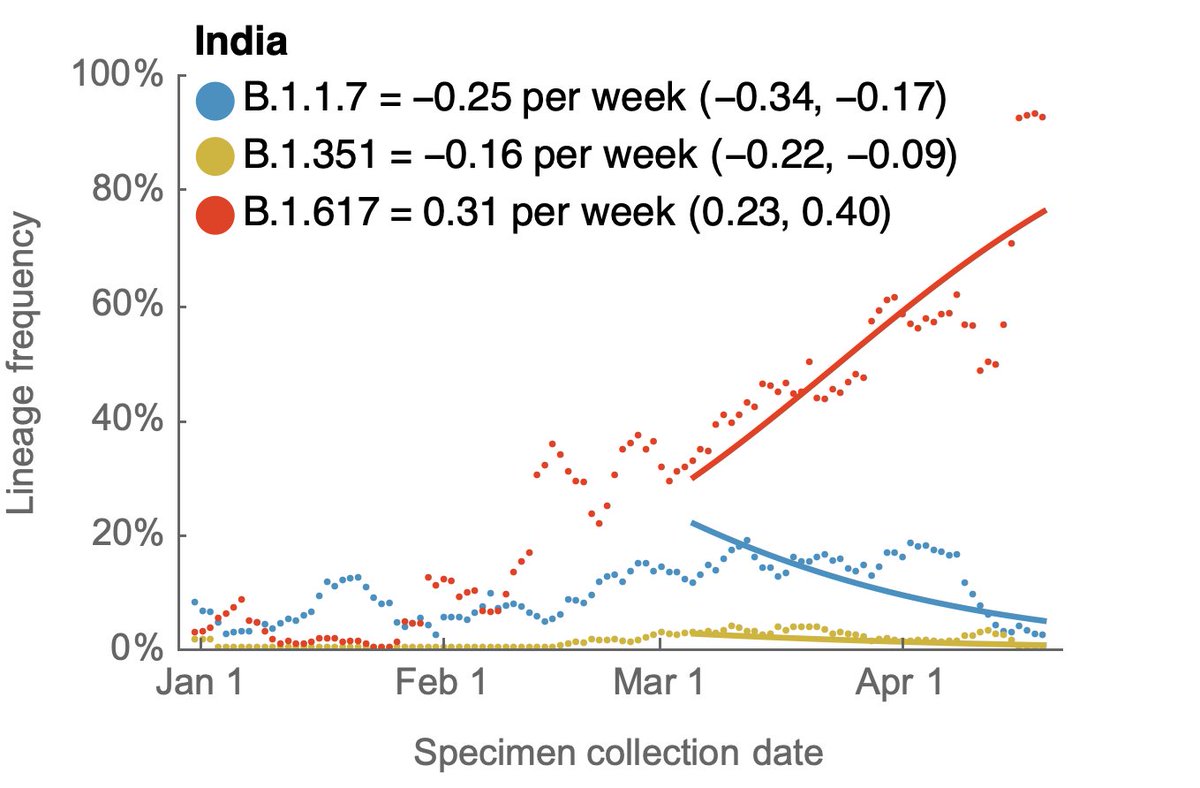
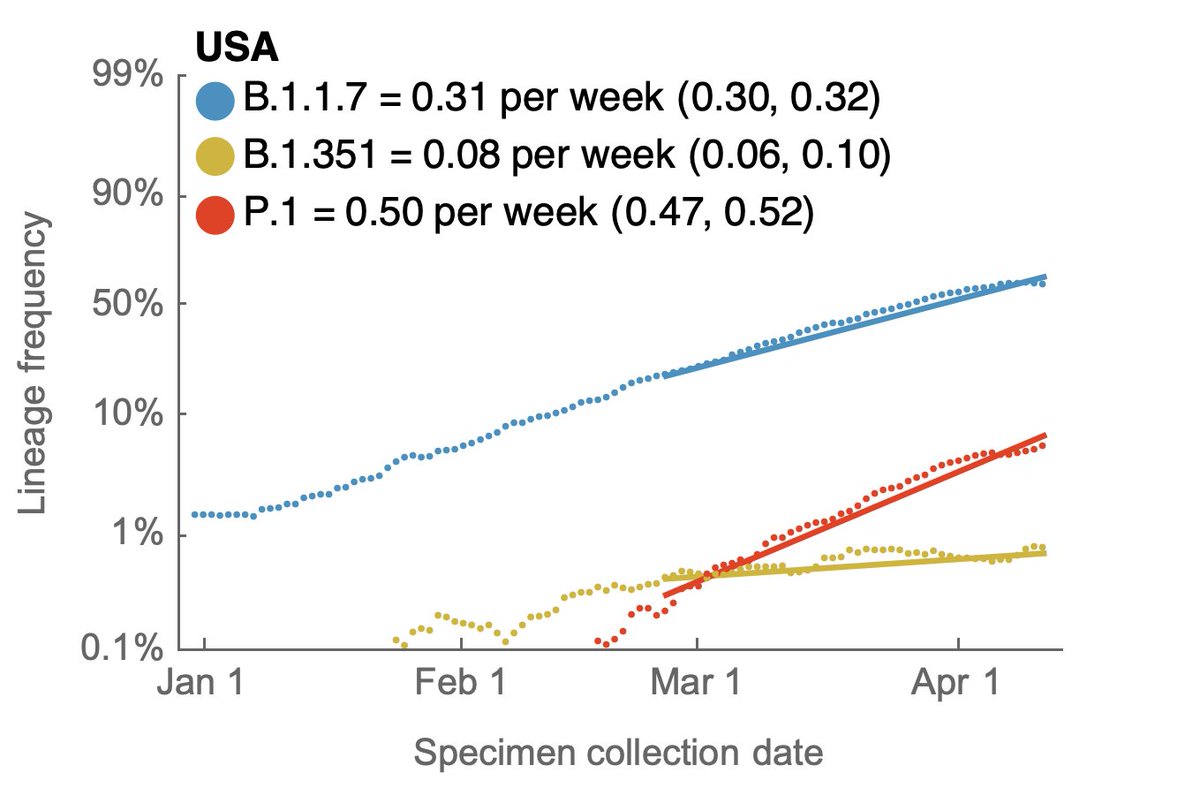
Looking across these nine states, the current ranking of logistic growth rate seems to be B.1.617 > P.1 > B.1.1.7 > B.1.526 > B.1.351. 6/10 

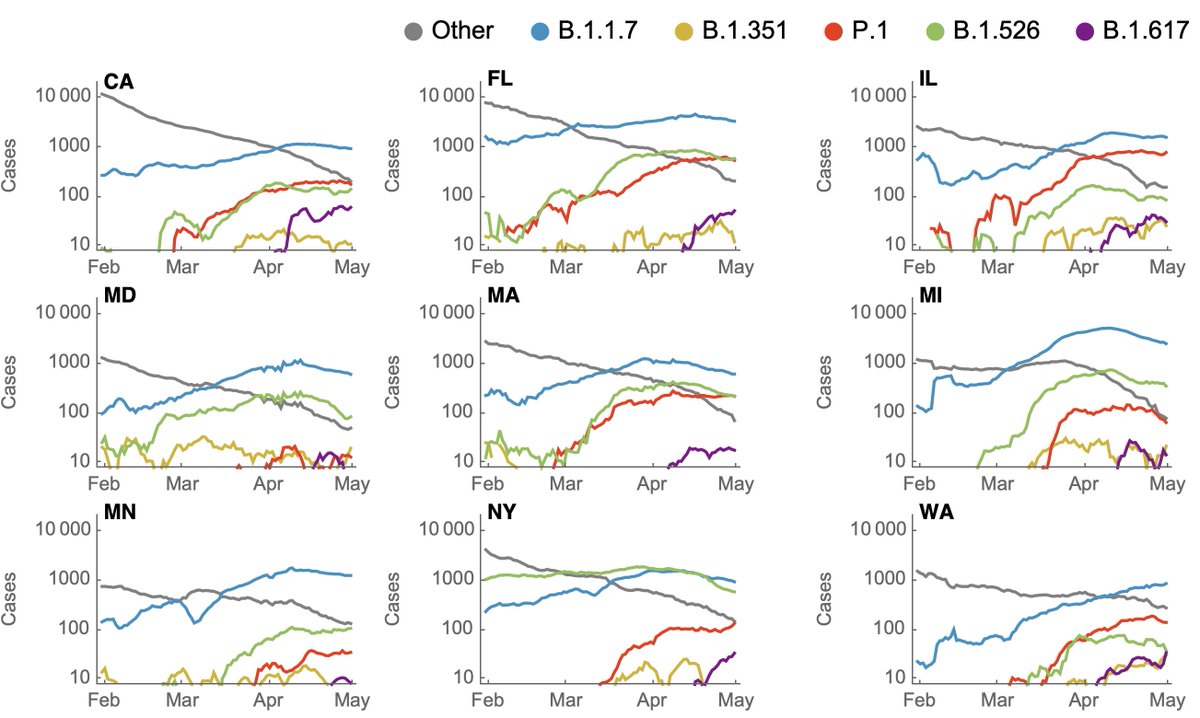
Although frequencies are useful to assess competitiveness of different variants, we're interested in case counts to assess whether a variant may be driving an epidemic. Here, I'm using genomic data to partition case counts as described previously. 7/10
https://twitter.com/trvrb/status/1389248456565297152
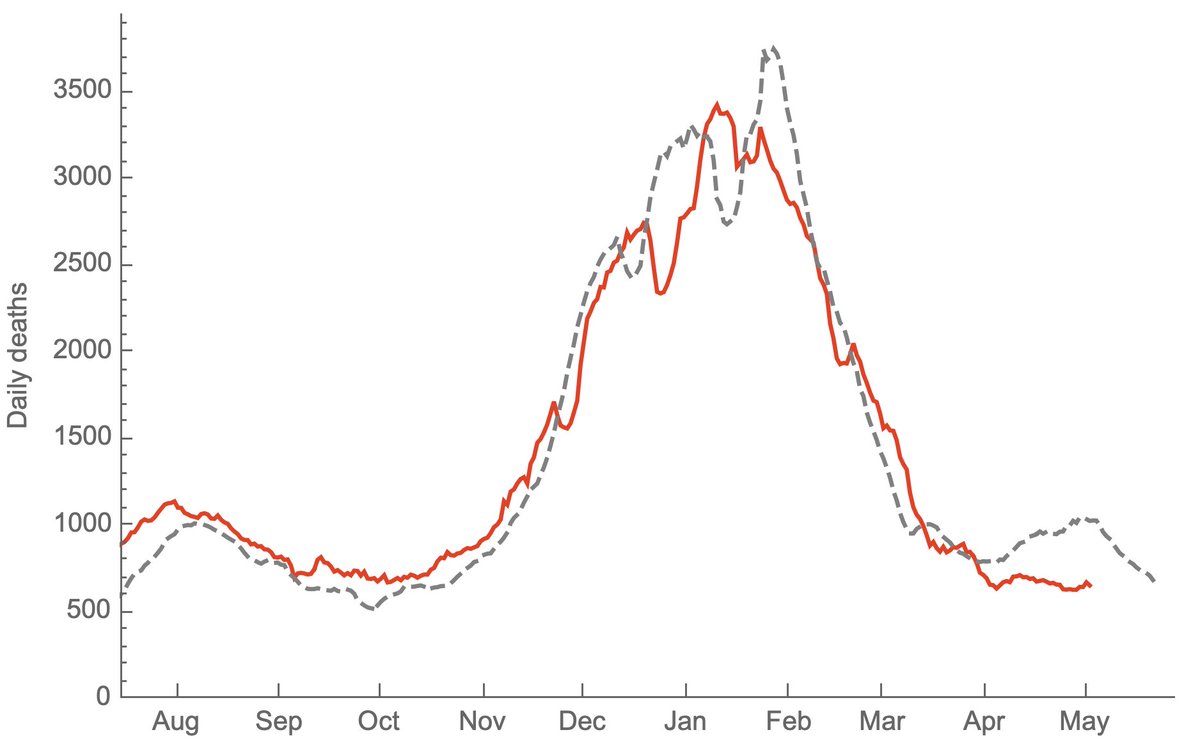
Doing so with this latest data gives the following picture where it's clear that non-variant viruses have been declining throughout the spring, while variant viruses have been responsible for multiple state-level epidemics (B.1.526 in NY, B.1.1.7 in MI, MD and MN). 8/10 

Because the genomic data is necessarily lagged, this is looking back to the beginning of May and the last 3 weeks of declines in cases are not included. However, even here, there are some encouraging trends of absolute growth of P.1 starting to level off. 9/10 
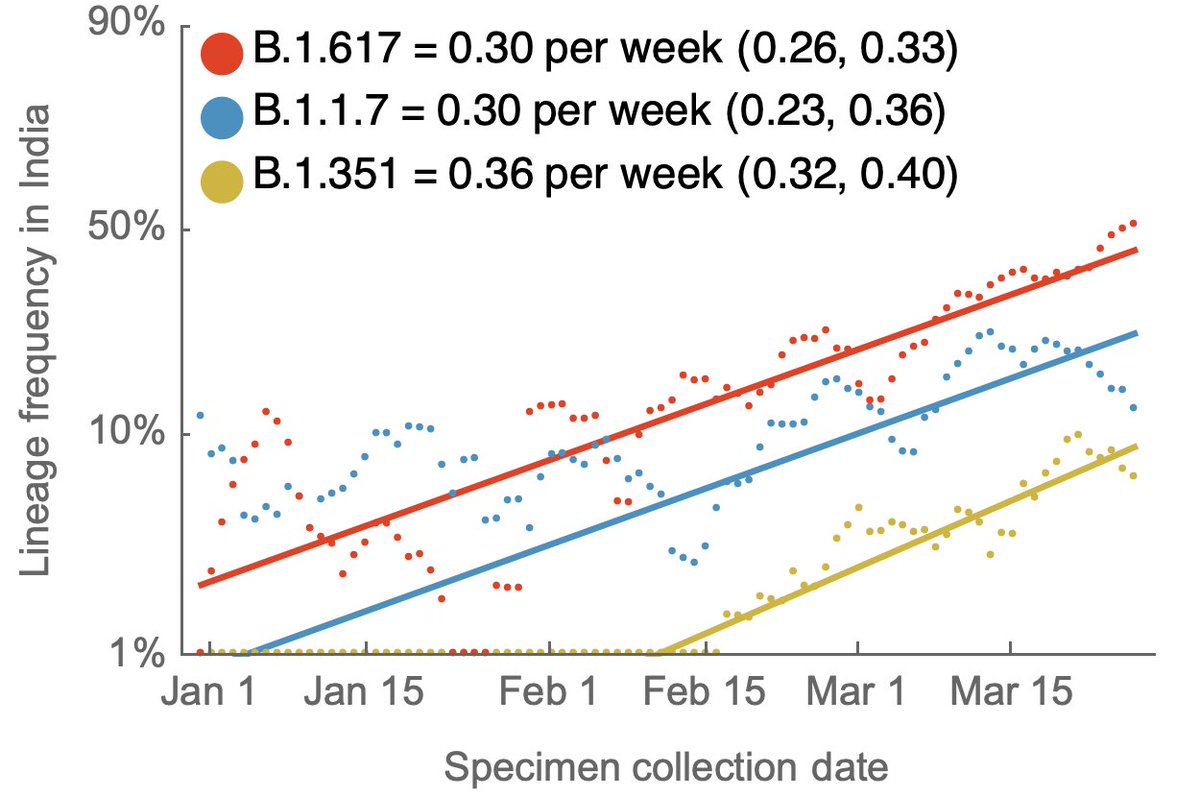
B.1.617 is, of course, worrisome and may gain ground quickly on other lineages, although its lack of significant immune escape makes me less worried about large-scale spread in the US. However, there is still a large unvaccinated population in which B.1.617 may drive cases. 10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh