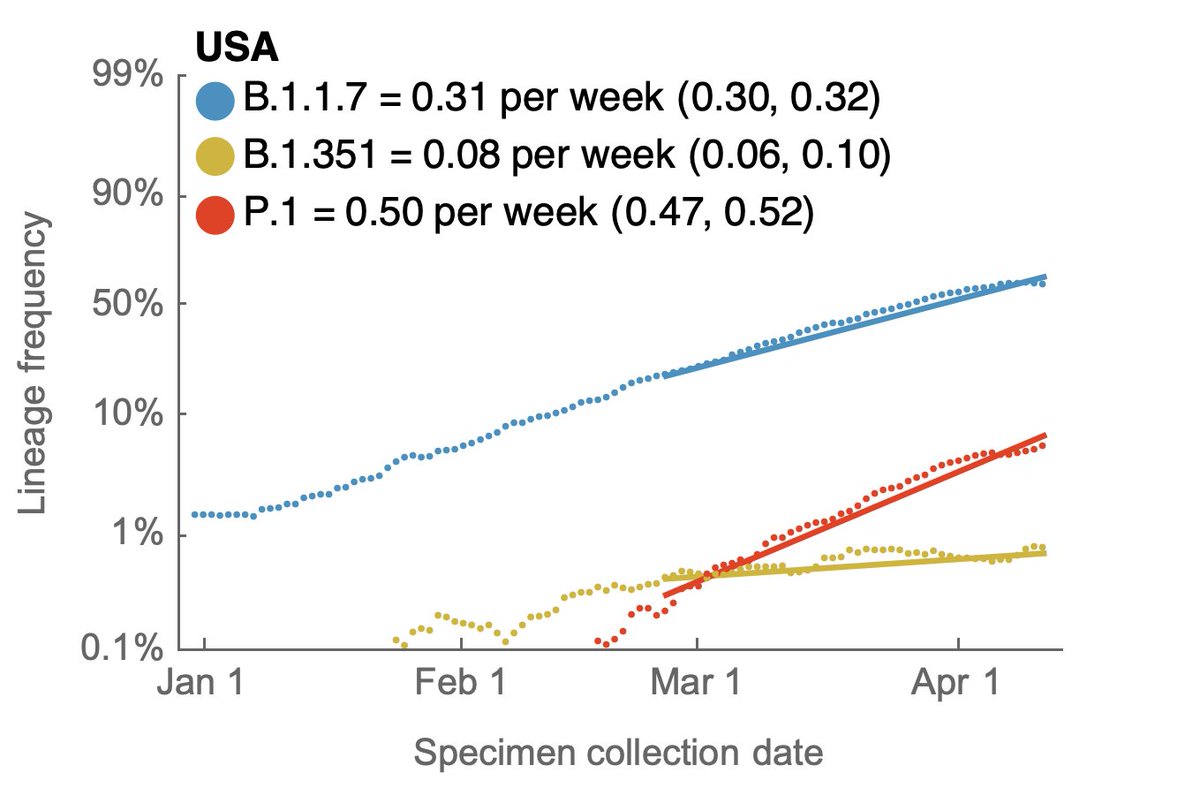
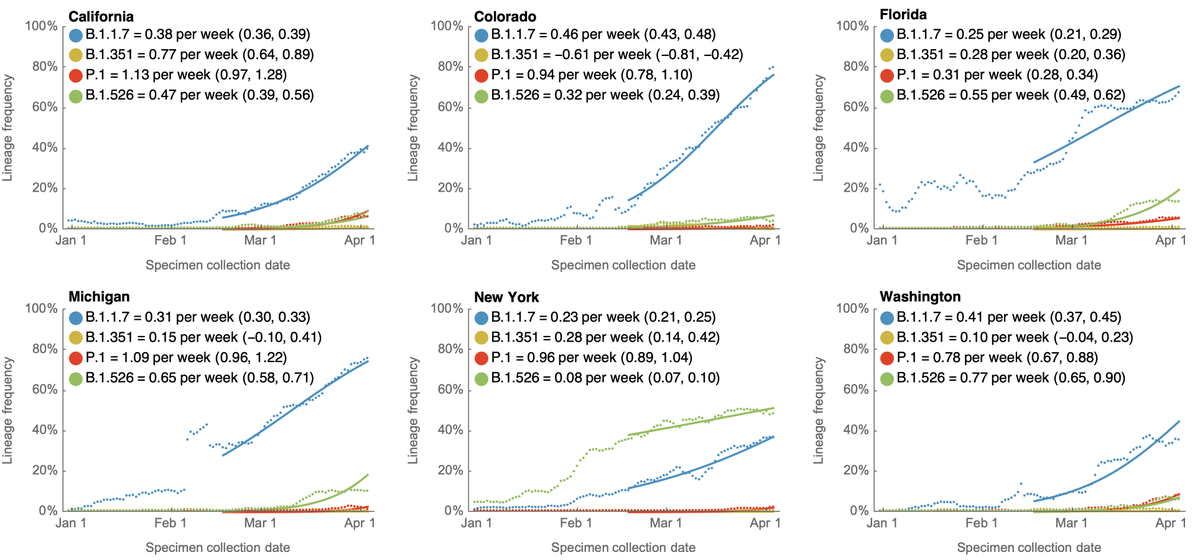
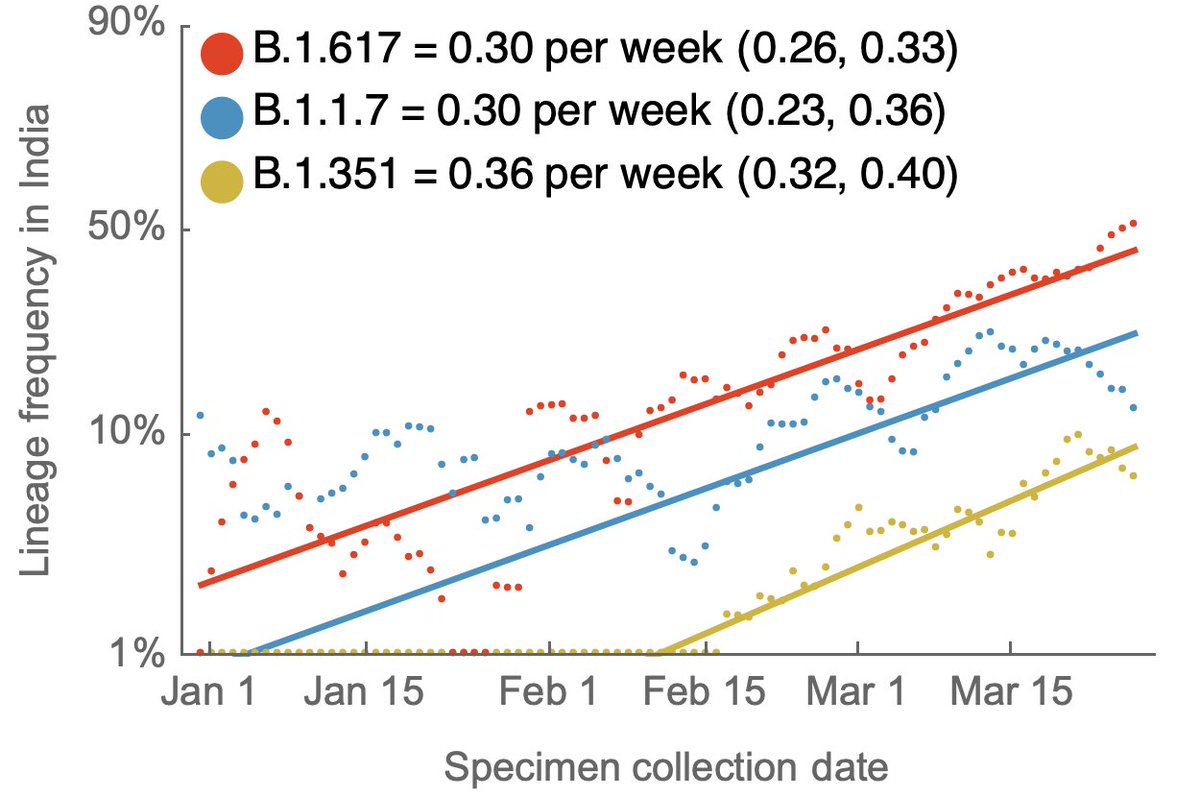
With the publication of the Science letter, the Overton window for discussion of "lab leak" hypothesis has shifted dramatically. We now have mainstream scientific opinions that largely range between "lab leak can be dismissed" and "both zoonosis and lab leak are viable". 1/8
I am in the both are plausible camp. The data (as it exists) is consistent with zoonosis, but it's also consistent with lab leak. Parsing the relative probabilities of the two depends on multiple lines of evidence and is necessarily assumption ridden. 2/8
https://twitter.com/trvrb/status/1333885880054910976
However, I think that there is a philosophical divide among scientists in how to assess hypotheses that perhaps explains some of the gap in opinion. Ie, is zoonosis the "null" hypothesis that we need significant evidence to reject or are we comparing two competing hypotheses? 3/8
Zoonoses occur constantly. We've had >26 Ebola outbreaks, 100s of MERS spillover events, abundant one-off human infections by avian influenza, etc... This is the prior many scientists are working from and suggests that zoonosis should be the null hypothesis. 4/8
On the other hand, of the recent influenza pandemics (H2N2 in 1957, H3N2 in 1968, H1N1 in 1977, H1N1 in 2009), one of the four (1977 H1N1) is conclusively a reintroduction into the human population via some lab intermediary. Lab accidents do happen. 5/8
If one takes this as naive prior of 1/4, one comes to a very different conclusion and would treat both zoonosis and lab leak as competing hypotheses for the (sparse) data. 6/8
I'm not sure how best to frame a prior in terms of origins of SARS-CoV-2, but I do agree that we'd benefit from a dispassionate evidence-based discourse on this issue. I also think that barring a major new datapoint, it's unlikely we'll come to definitive conclusions. 7/8
Regardless of COVID-19 origins, we should treat zoonosis and lab escape as potential pandemic risks and set up structures to mitigate these risks. On the lab side, this includes review of biosafety protocols and strong "no fault" reporting of laboratory acquired infections. 8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh