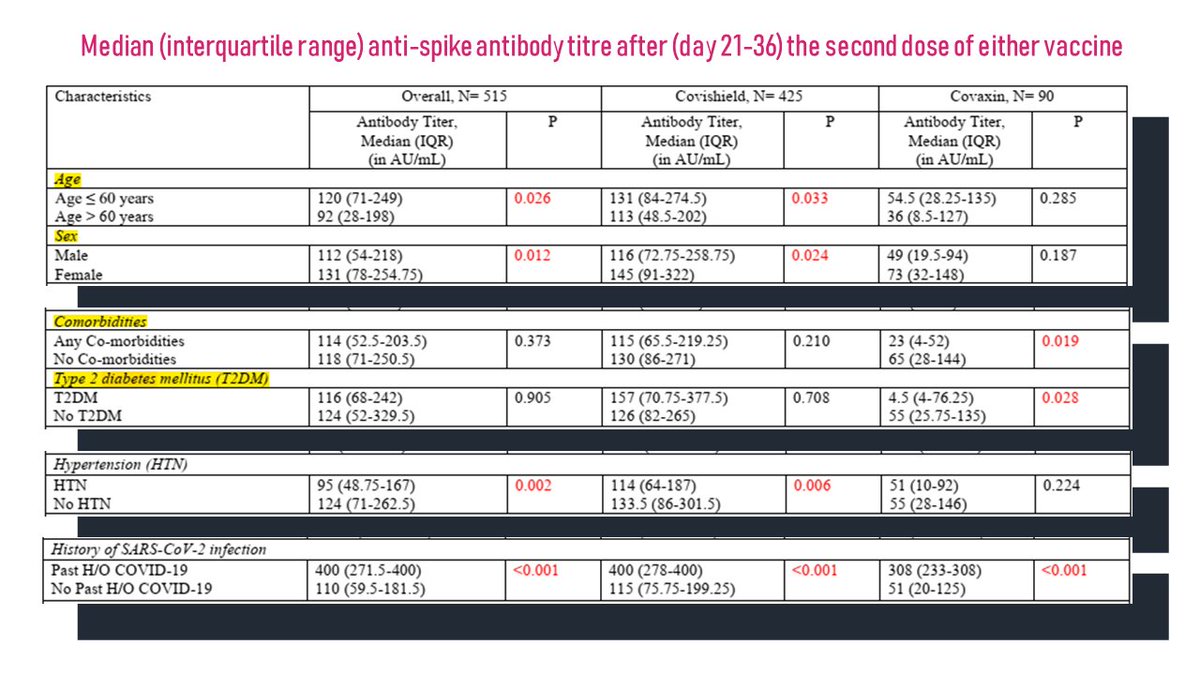
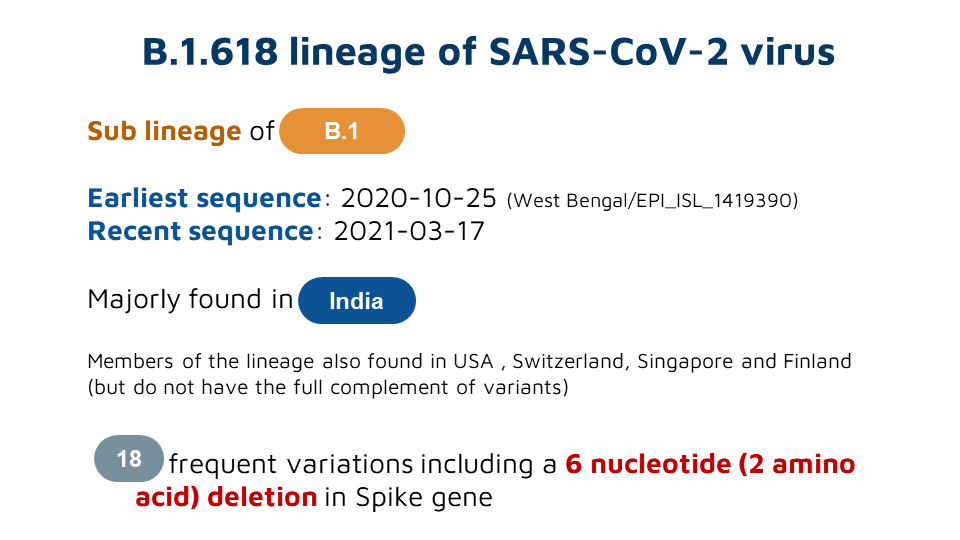
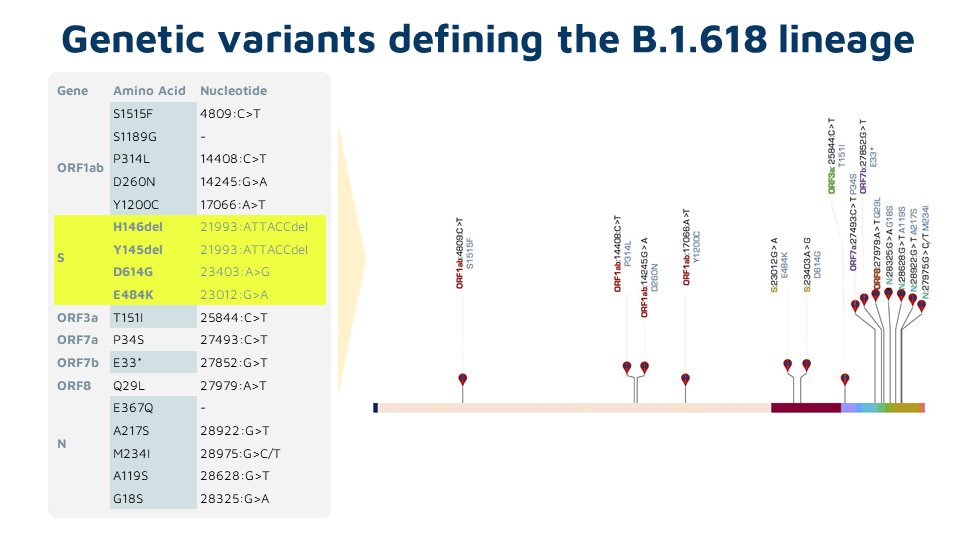
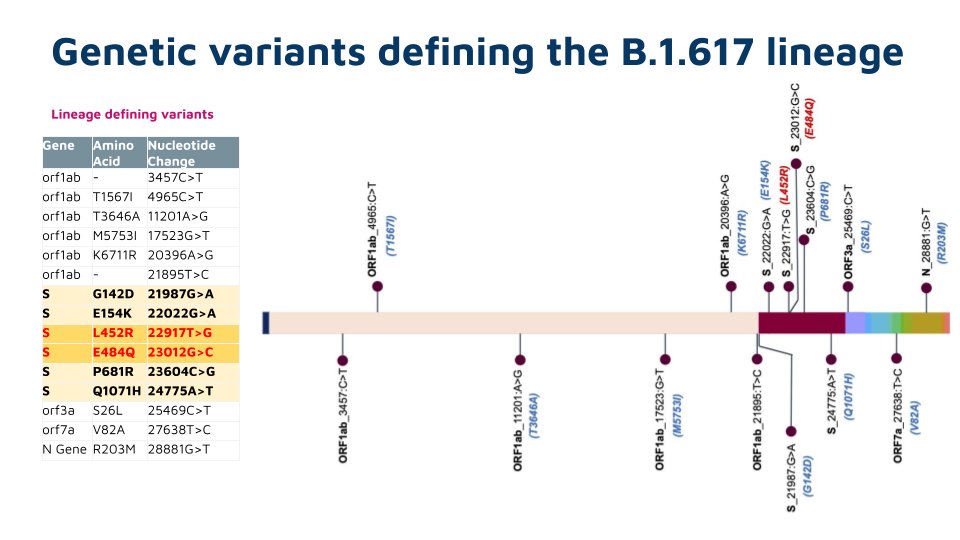
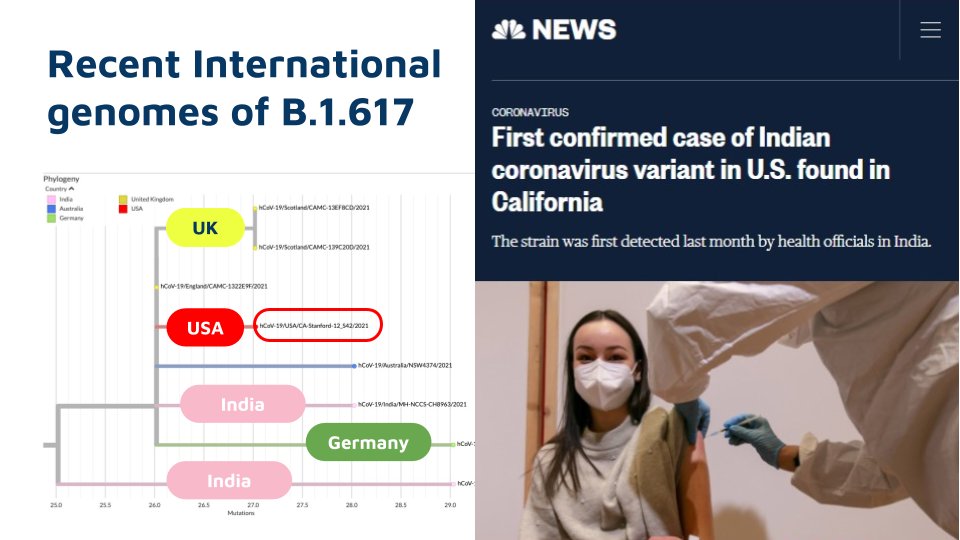
As delta continues to evolve acquiring new mutations, there is a lot if interest in understanding these mutations. This tweetorial summarizes the emerging lineages of delta, otherwise named delta+
Data and analysis : @bani_jolly and @mercy_rophina
Data and analysis : @bani_jolly and @mercy_rophina

Understanding this continued evolution is of great importance in mapping the evolutionary landscape of emerging variants. Largely the virus has tried to optmise for transmission and immune escape by step-wise acquisition of new mutations. 

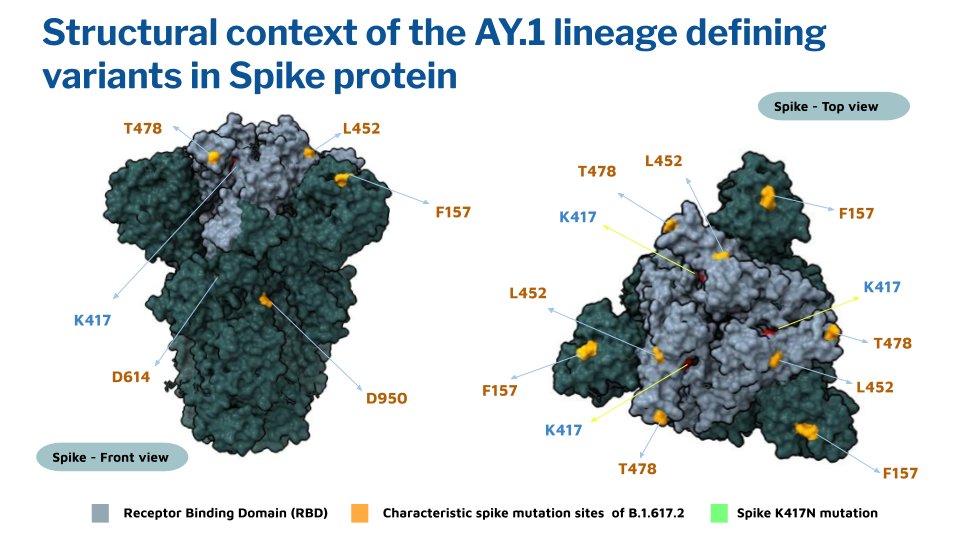
One of the emerging variants is B.1.617.2.1 also known as AY.1 characterized by the acquisition of K417N mutation.
Thanks @bani_jolly for pointing out the mutation and lineage assignment.
Thanks @bani_jolly for pointing out the mutation and lineage assignment.

The context of the K417N mutation is shown in figure below. The mutation maps to the receptor binding domain and has also been associated with immune escape
Ref: clingen.igib.res.in/esc
Schema: @mercy_rophina
Ref: clingen.igib.res.in/esc
Schema: @mercy_rophina
A number of genomes are now available for the lineage AY.1 / B.1.617.2.1 from across the world. The sequences are mostly from Europe, Asia and America. The travel histories are not readily available to make assumptions. 

One important point to consider regarding K417N is evidence suggesting resistance to mAbs Casirivimab and Imdevimab. The mAb cocktail incidentally has received an EUA from @CDSCO_INDIA_INF in India.
The variant freq for K417N is not much in India at this point in time
The variant freq for K417N is not much in India at this point in time

The recent @PHE_uk report also has a mention of the variant in 36 cases in England.
assets.publishing.service.gov.uk/government/upl…
2 of the 36 were possible vaccine breakthrough infections
assets.publishing.service.gov.uk/government/upl…
2 of the 36 were possible vaccine breakthrough infections

A dedicated nextstrain build for K417N is also now available .
@firefoxx66
@firefoxx66
https://twitter.com/firefoxx66/status/1402575410785837056?s=20
There is a very informative thread on the evolution and phylogenetic context of AY.1 by @bani_jolly 👇
https://twitter.com/bani_jolly/status/1404382022714331138?s=20
• • •
Missing some Tweet in this thread? You can try to
force a refresh