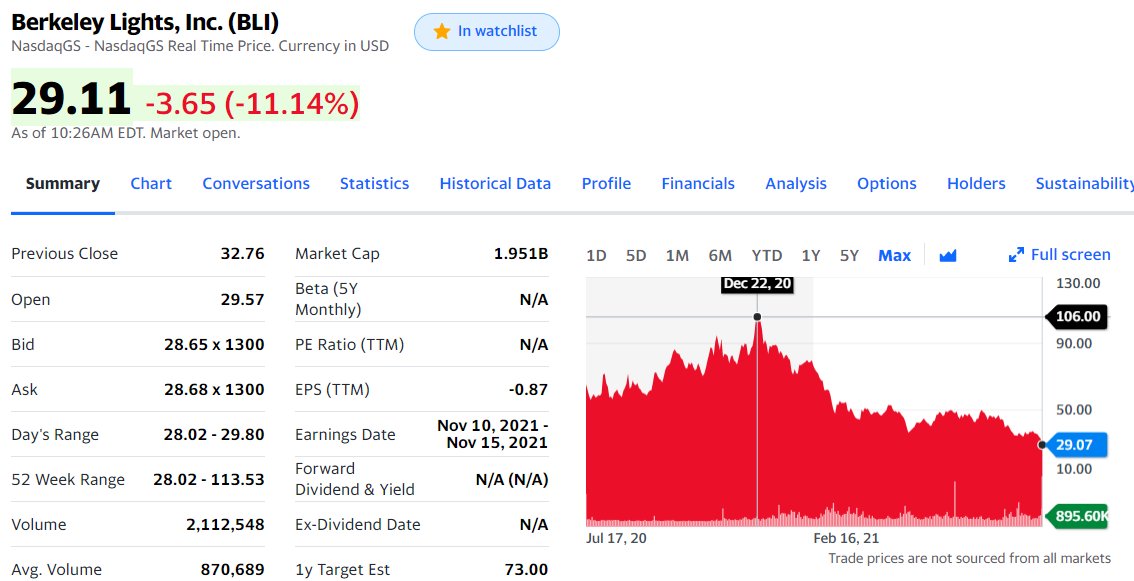
(1) Possible buyout of #ARM by #NVIDIA does have an effect on the #Bioinformatics field: many applications now are deployable on CPU/GPUs with #ARM and/or #NVIDIA chips on them. Some recent examples are:
(a) the Oxford @nanopore MinION Mk1c device, which originally was specced at Jetson TX2 ARM+Pascal GPU accelerators (ARM processor 6 cores, 256 Core GPU), 8 GB RAM (may have changed since then.
(b) #Alphafold2 is another recent example that, although it can work without an #NVIDIA #GPU, but works much faster and efficiently on it. It can predict the folding of proteins in the 1000-1500aa length range on a GPU card with 16Gb of memory.
(2) Possible buyout of @GrailBio by @illumina: this is, in theory, going forward but it could be legally challenged by US and EU (maybe other) regulators. This has less of a direct impact on anyone: Grail's Galleri is available in the US and being piloted at NHS England.
And regardless of what the merger or no-merger ends up looking like, it's difficult to see it will impact the current availability of the Galleri test. But it could impact the availability of other #LiquidBiopsy #Epigenomic profiling #CancerScreening tests out there.
Given that @illumina wants to quickly vertically integrate into the largest and potentially most profitable Dx market for #NGS #shortreads, they have a limited window of time to combine the technological lead of GrailBio into the deployment lead they think the merger will have.
If one assumes that Grail's Galleri is the leading technology that has already been approved by the FDA in #LiquidBiopsy multi #CancerScreening, then the only resort for Exact Sciences and other bigger players is to deploy their assays more widely and affordably than Grail.
Given that @illumina keeps a roughly 80-90% margin on their largest #NGS sequencers, they would be able, in theory, to internally lower the cost, and externally the price, of Grail's Galleri assay for as long as it takes to flood the Dx market and obliterate the competition.
So it could be argued that it's a newcomer, Grail, backed by an instruments company, Illumina, that wants to take over the market of the current oligopolies, Exact Sciences with Cologuard, maybe Guardant Health, but I wouldn't call them oligopolies.
In single-payer health systems, like the NHS, there is an entity dedicated to evaluating the pros/cons of each assay, and what will it mean to downstream medical care to be able to identify a certain % of Stage I cancers.
In an ideal world, one would get annually tested with a #LiquidBiopsy #Epigenomic profiling assay, then only if something doesn't look right, then be referred to further treatment. In cancer, early diagnosis could mean many things: they may MRI the patient on a full-body scan,
or they make go straight to surgery to look closely into the tissue/organ that the Dx assay has identified, maybe a benign-looking growth is resected, or maybe nothing is resected. But that's a much better outcome than letting the cancer move into Stage II, III or IV.
Also important to note that #epigenomic profiling of #CancerScreening works on the basis of altered cell states at the tumor microenvironment, and it can identify, e.g. for colorectal cancer, benign polyps that may not even test positive for a panel of cancer SNVs.
... identified
One argument against the merger is that we have a healthy ecosystem now that #ARM is an independent entity, supplying designs for CPUs and GPUs to a variety of other large/medium/small entities. Likewise, #NVIDIA is a big player in #GPU design and production but isn't in #CPUs.
So with that logic in mind, what we've experienced in the last decade or so is an example of a well-balanced ecosystem and market, bearing fruits in a great variety of smartphones, tablets, and other consumer devices. No one has taken a dominating position, even when tried.
For example, #Apple made a couple of big moves in attempting to take a technological lead in smartphone chips, but those were mostly not as successful as they wanted. Arguably, the top-end CPU/GPUs for smartphones are still designed by #ARM or heavily influenced by the ...
... design phylosophy that #ARM adopted in the last decade, with the Cambrian explosion of smartphone/tablet/wristwatch devices.
So what's the risk of #ARM and #NVIDIA becoming a single entity? We could look at it historically to see if we learn anything from the recent past.
Back when smartphones were becoming a thing, there was a transition from the relatively simple, sometimes coined "dumb" phones ...
Back when smartphones were becoming a thing, there was a transition from the relatively simple, sometimes coined "dumb" phones ...
... towards what we now know as smartphones. This transition involved upgrading the chips/CPUs/GPUs into bigger more powerful computational devices, but it was done almost exclusively from a base of dumb phones that had a very good power usage profile.
In hindsight, it could have happened the other way around: larger laptops becoming smaller by making their CPUs/GPUs more energy efficient, to the point of intersection with the dumbphones battery life. And there were powerful entities attempting this route at the time:
Intel, Microsoft, Sony, Toshiba, even IBM wanted to turn their power-efficient laptop motherboards/chips into powerful smartphones that would still last a working day in people's hands. But they failed: even though they were very large entities with a lot of influence over ...
... the market, they couldn't ship products that would compete with the other route, from dumbphones to smartphones.
And who ended up as winners in this technology race? Companies like Apple, but also Samsung, Xiaomi, LG, Huawei, Oppo, and earlier on also ZTE, HTC, Motorola, Nokia, etc. en.wikipedia.org/wiki/List_of_b…
Many of these companies weren't big and powerful when they entered the smartphone market. But they managed to hire good engineers, they paid some money to #ARM to have access to their #CPU and #GPU designs and didn't have any other constraints beyond that.
What would have happened at that point if #ARM had been gobbled up and reduced the freedom to operate (#FTO) of all the nascent players in the smartphone market? They would have limited the diversity of the ecosystem, and we would have ended with fewer/lesser choices/products.
So there is the argument that #NVIDIA gobbling up #ARM could be deemed a relevant antitrust scenario for the regulators in the US/EU and elsewhere.
How could that affect #Genomics and #Bioinformatics? In some ways, we already see that the lack of diversity in the #GPU market is stifling innovation: when #NVIDIA came up with #CUDA, their cards were ubiquitous enough that they started to be adopted in #Bioinformatics (GPGPUs).
At that time, #ATI (later #AMD) and a few others came up with a #CUDA equivalent called #OpenCL en.wikipedia.org/wiki/OpenCL. That move could have had enough momentum for #NVIDIA to ditch #CUDA and join the #OpenCL consortium, but they didn't.
What that chiasm meant a few years later is that each software project wanting to accelerate their code had to adopt #CUDA *and* then re-implement their code hooks for #OpenCL if they wanted both to be compatible with their code/libraries.
Many software projects decided the order in which they would adopt these based on install-base, which meant #NVIDIA #CUDA was quickly adopted, but #OpenCL and related (e.g. SYCL) weren't adopted as much.
For example, #TensorFlow is written in Python, C++ and #CUDA, and they don't have the same level of support for #OpenCL or related. There is even support for #TPU platforms, sponsored by Google, the only company that owns them. en.wikipedia.org/wiki/Tensor_Pr…
So the General Purpose GPU ecosystem (#GPGPU) is a working system, but compared to smartphones, it has been less diverse and fruitful than it could have been. This means that when someone is wanting to write a clever piece of code for #Bioinformatics, e.g. Alphafold2, there is...
less choice available to them to deploy that code in a combination of CPUs/GPGPUs/TPUs, etc. that will make it run fast and efficiently. This combined with the fact that #cryptomining has made GPU cards a lot less available for purchase means that it's not an easy field to be.
Moving forward, there will be many more applications in #Genomics and #Bioinformatics that will benefit from power-efficient portable #ARM devices, and Oxford @nanopore is an almost textbook example of a future that's already with us with their MinION Mk1c.
Oxford @nanopore MinION Mk1c has a Max rated power of 60W and Max rated current of 10A, which compares very well with current smartphones/tablets. It also looks like a tablet, so even for the IT averse it won't feel intimidating to use, as they are used to tablet-looking gizmos. 
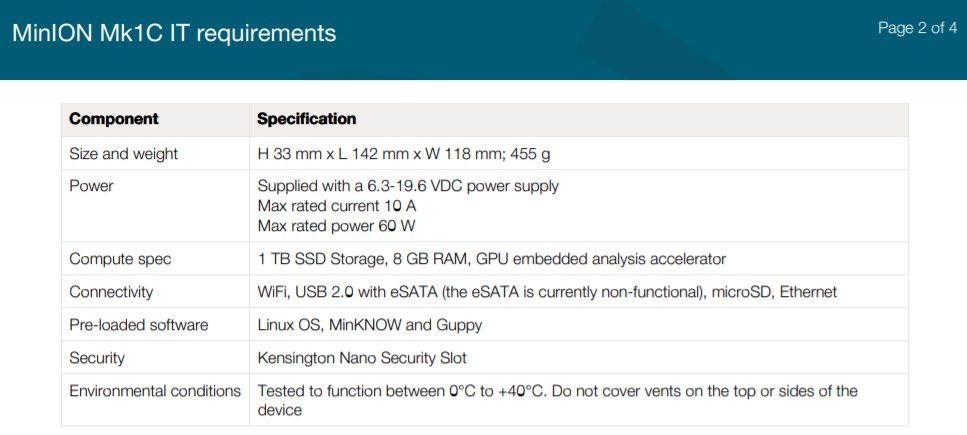
Given improvements in DNA sample prep ancillaries, and as the price of #NGS keeps going down, we are getting closer and closer to ubiquitous DNA/RNA-based assays that can be as affordable as an off-the-shelf test.
But these and other examples require a diverse and thriving ecosystem in the smart portable computing field to make the best, most disruptive #Genomics devices possible.
So let's hope that for both antitrust headlines, the #NVIDIA / #ARM deal and the #Illumina / #GrailBio deal, we can see a resolution that means the technology will keep growing, be quickly and widely adopted in the right places, and keep as much freedom-to-operate for as many.
@d2unroll unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh