Hello from Peru. Recently, there's been some buzz about Lambda, Mu, and variants coming out of South America. Here's a summary of the variant landscape in the region where Delta is yet to dominate (but will soon), as we prepare for a new wave of cases in the following weeks.
1/n


1/n


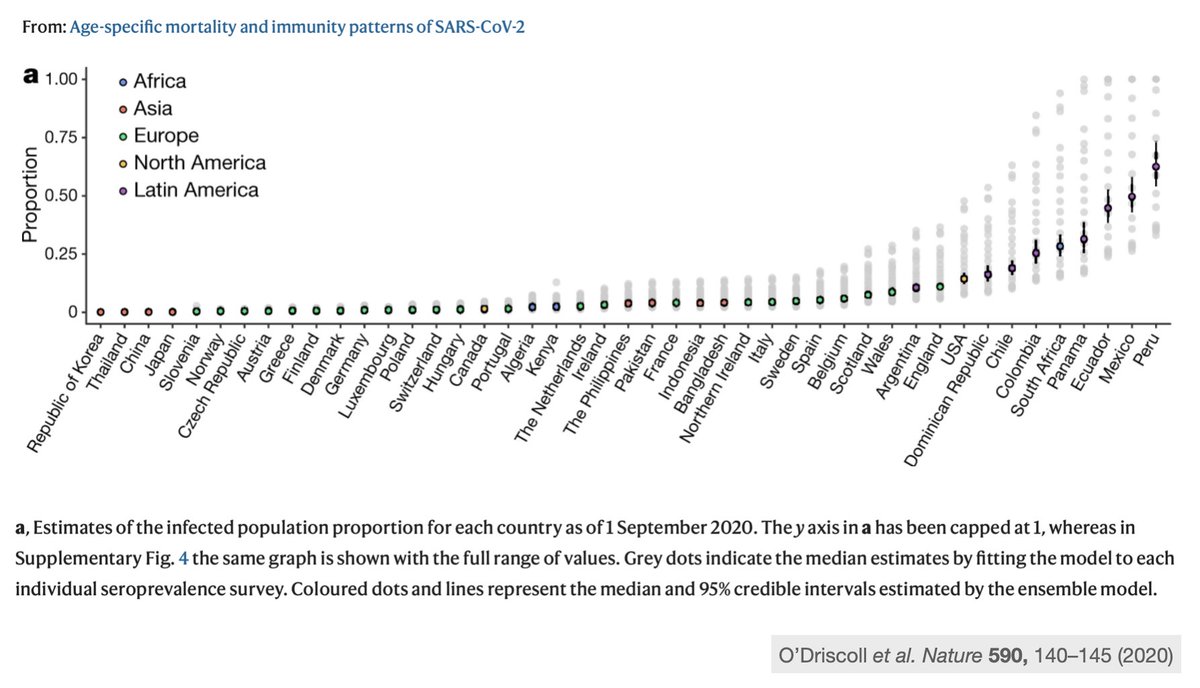
Let me first bring your attention to Latin America, an epicenter for COVID-19 since the start of the pandemic. We have around 8% of the world population (660M) yet accumulate 25+% (1.4M) of COVID deaths.
ft.com/content/a2901c…
2/
ft.com/content/a2901c…
2/

While much attention has focused on Brazil due to the magnitude of its epidemic (584k+ COVID deaths, pop. 210M+), relative to pop size, many countries in the region have had similar or worse epidemics.
3/



3/




Looking at excess deaths relative to previous years, many Latam countries appear at the top. ft.com/content/a2901c…
4/

4/


So, we have had very high levels of transmission and it's not surprising that novel variants beyond P.1 would emerge in the region, usually in the presence of high seroprevalence from the initial wave of cases in 2020.
go.nature.com/3DXhrJR
bit.ly/3yVcGww
5/
go.nature.com/3DXhrJR
bit.ly/3yVcGww
5/
South America has a very limited capacity for genomic surveillance: We contribute less than 2% of sequences on GISAID (65k by September 8), which represent less than 0.5% of total cases for most countries. We are also very slow to upload sequences to GISAID/Genbank.
6/
6/

More on the global disparities in genomic surveillance in this post from @AndersonBrito_
7/
https://twitter.com/AndersonBrito_/status/1431235752944340992?s=20
7/
P.1 / Gamma is already known to everyone. Its emergence from Manaus and its spread have been described in detail by our colleagues at CADDE, Fiocruz, and others in Brazil.
science.org/doi/full/10.11…
nature.com/articles/s4159…
8/
science.org/doi/full/10.11…
nature.com/articles/s4159…
8/

Gamma has now generated multiple sublineages, has been exported to 80+ countries, and is arguably the most successful variant in the region so far. outbreak.info/situation-repo…
cov-lineages.org/lineage.html?l…
9/

cov-lineages.org/lineage.html?l…
9/


Elsewhere in Brazil, P.2 / Zeta evolved independently from P.1 and had S:E484K but not N501Y or K417N. It peaked around December and has been exported to 40+ countries but has since been replaced by P.1 in most of Brazil.
journals.asm.org/doi/10.1128/JV…
10/


journals.asm.org/doi/10.1128/JV…
10/

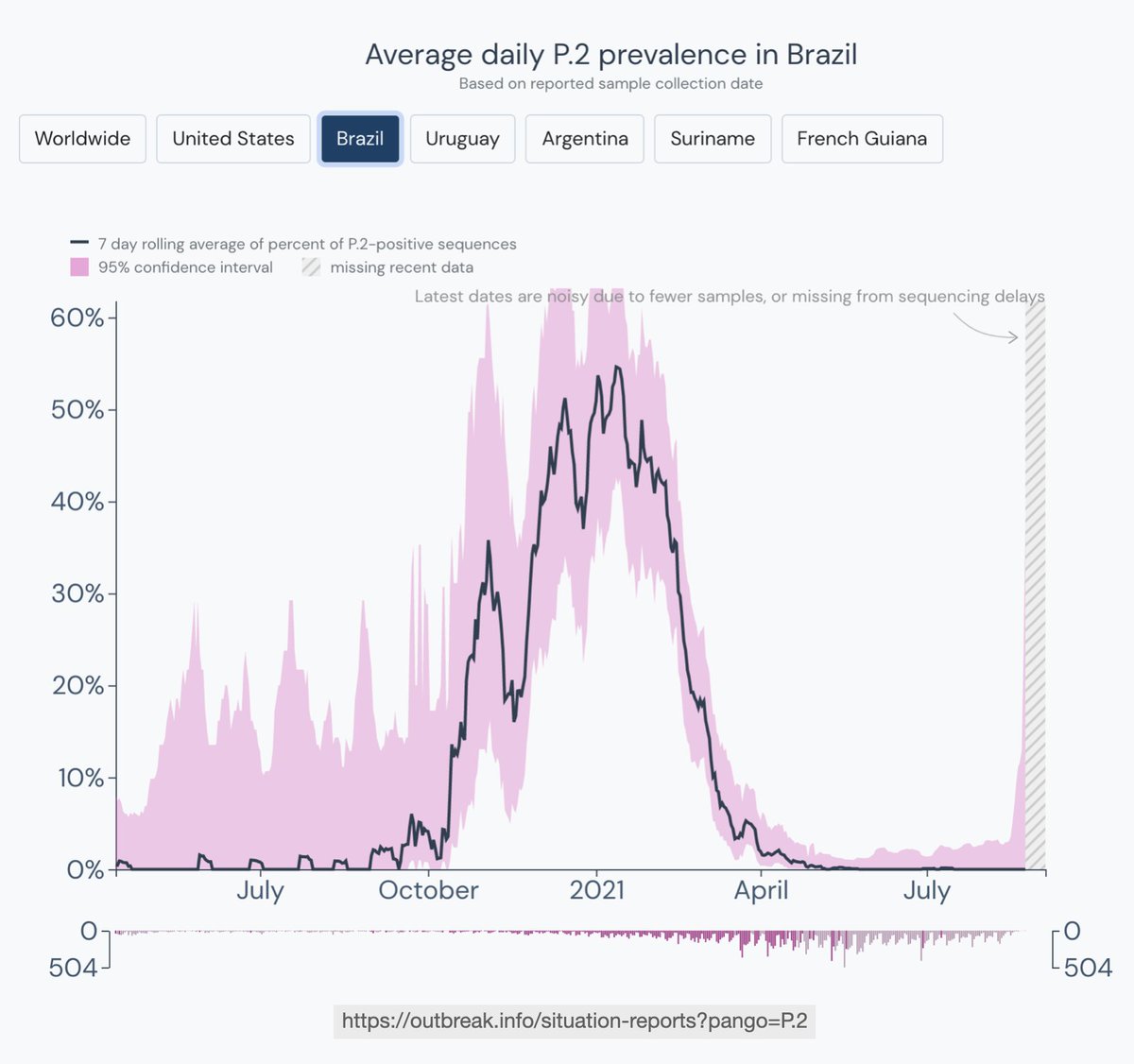

Moving to Uruguay, which contained the virus for most of 2020. Cases rose in November with P.6, a B.1.1.28 sublineage with S:Q675H+Q677H. P.6 was replaced by P.1 by April, coinciding with a sharp rise in cases and deaths.
go.nature.com/3jTTlrm
bit.ly/3tunQHt
11/


go.nature.com/3jTTlrm
bit.ly/3tunQHt
11/


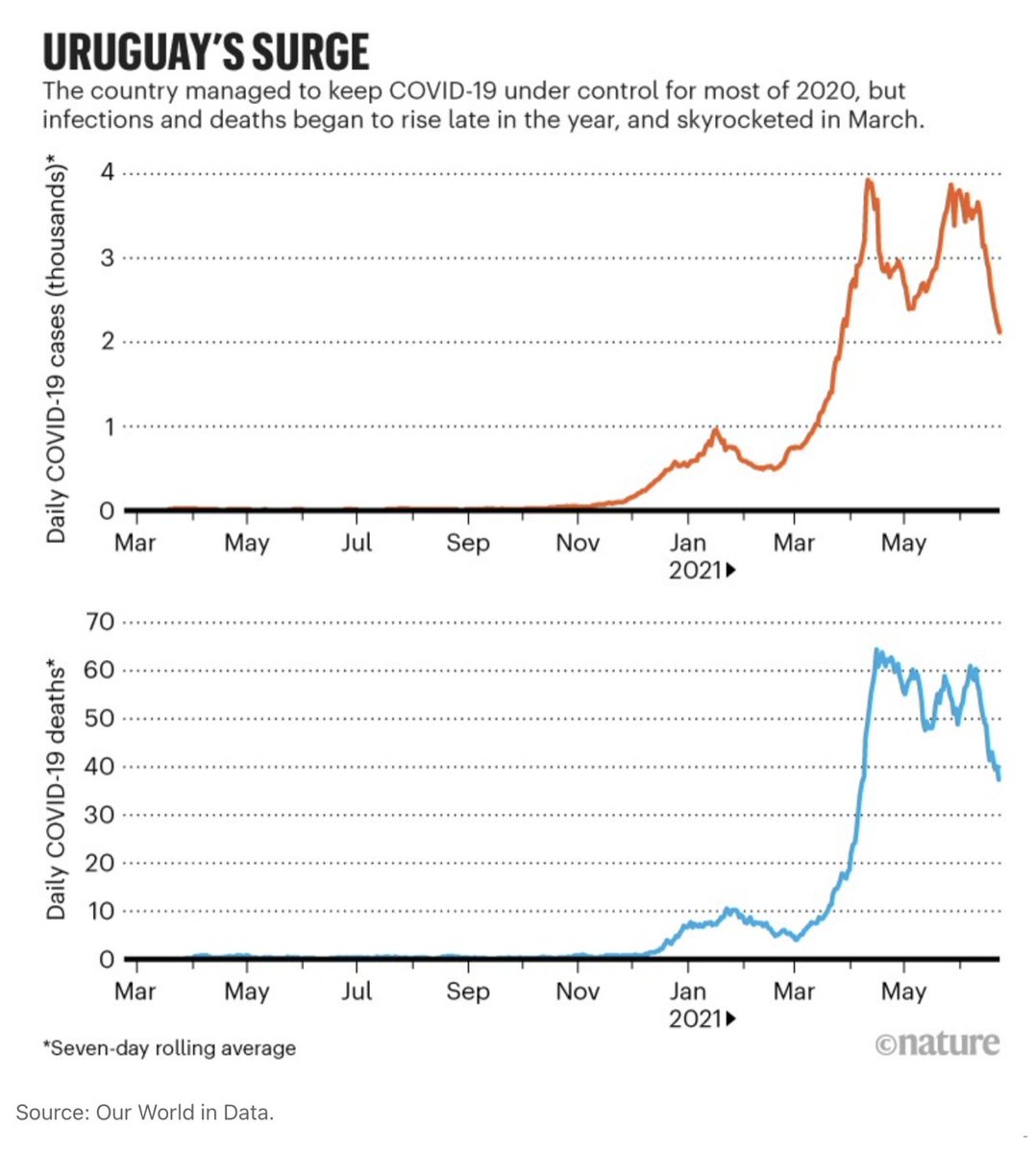
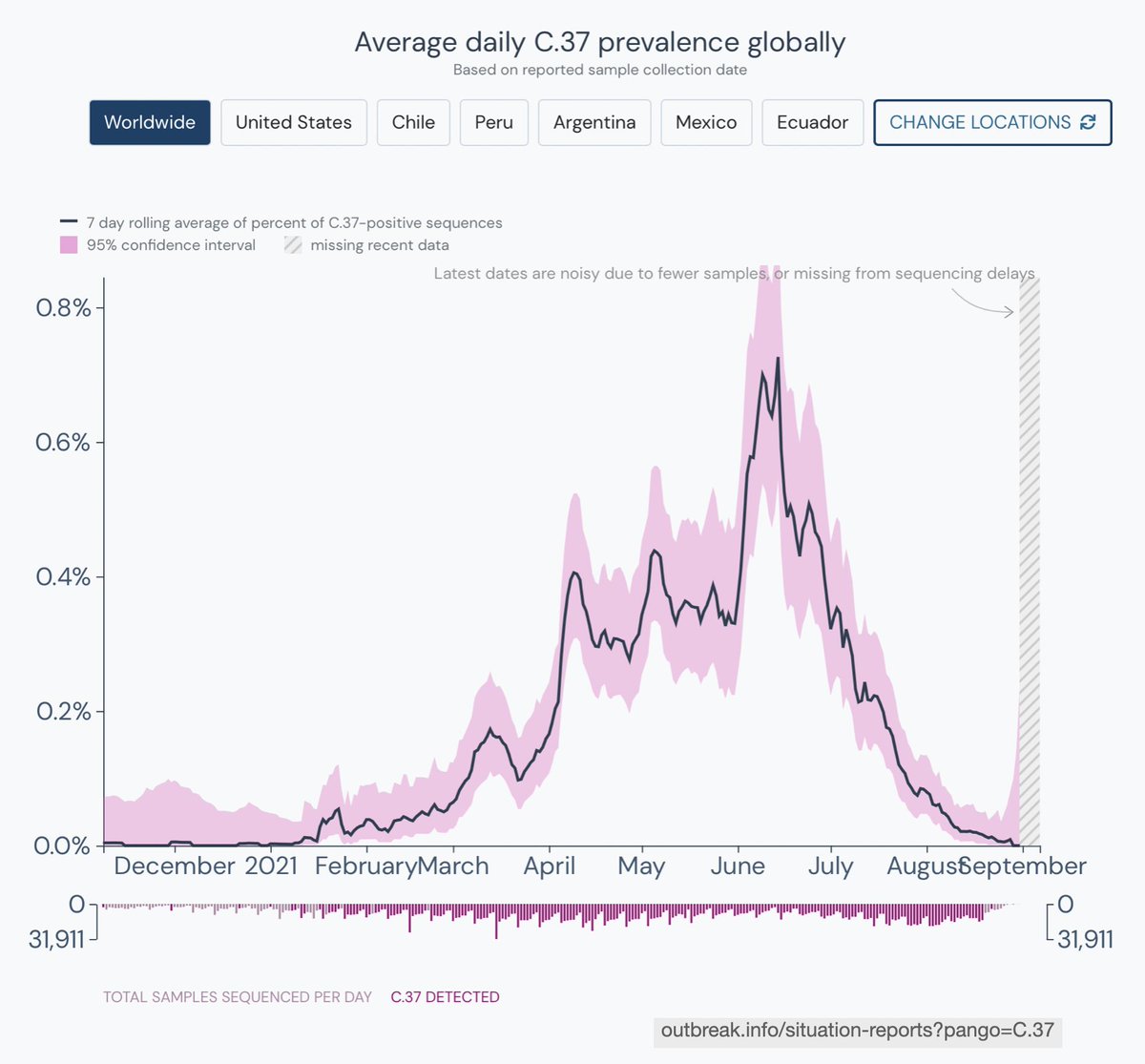
Now, on to C.37 / Lambda. I will explain more about it since I am most familiar with this story.
medrxiv.org/content/10.110…
12/
medrxiv.org/content/10.110…
12/
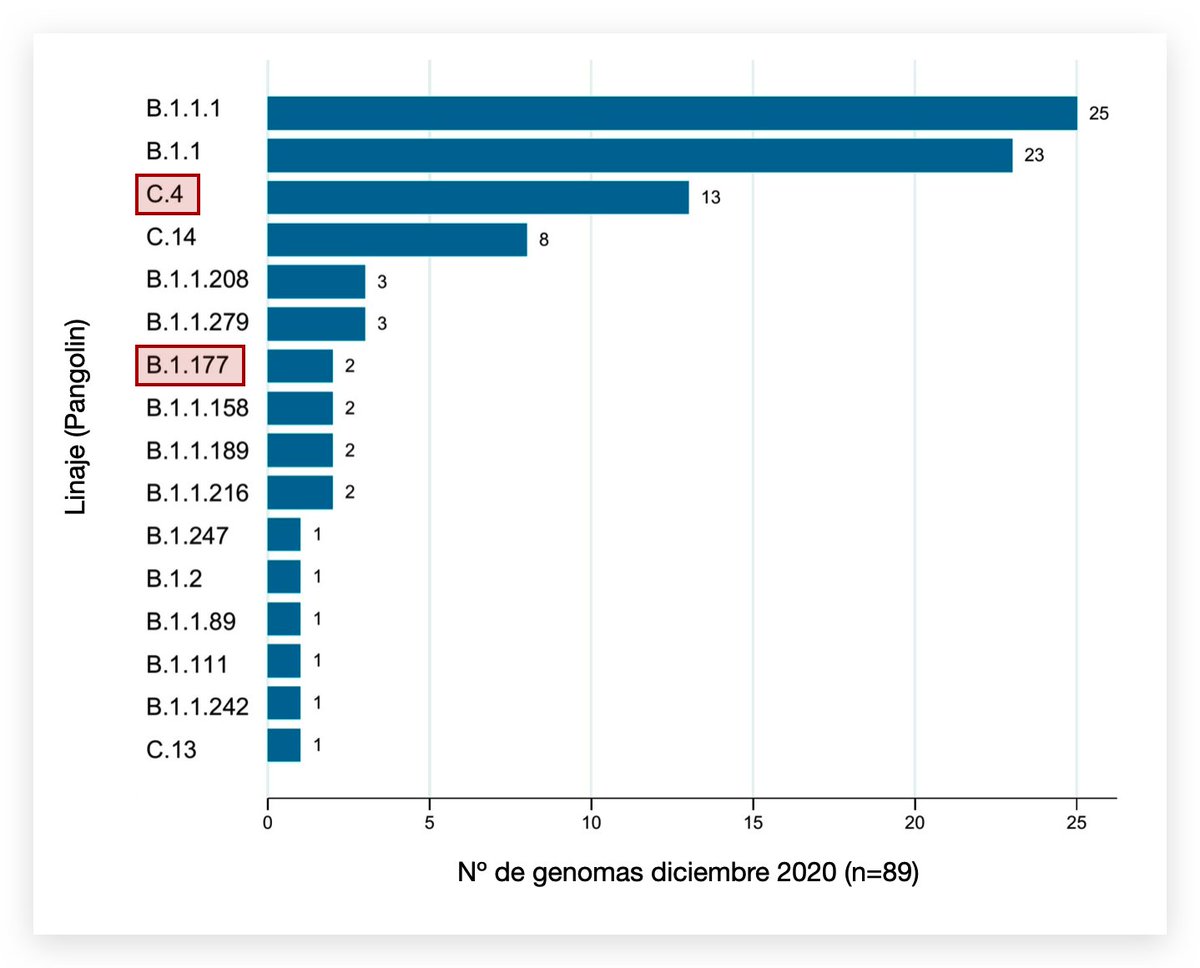
C.37 evolved from B.1.1.1, a lineage imported from Europe early in the pandemic. By late 2020, B.1.1.1 accounted for 20%+ of sequenced genomes from Peru.
14/
14/

Lambda has its unique constellation of mutations in the Spike gene. Particularly interesting at first: Δ247-253 in NTD, and L452Q (similar to L452R in Delta) + F490S in RBD. It also shares a convergent deletion in ORF1a:3675-3677 with Alpha, Beta, Gamma, Eta, and Iota.
15/
15/

We first noticed it in Lima in late December (<1% of cases then). By April, at the peak of Peru's second wave, it was already 80% of all sequenced cases.
16/
16/

By the time we first reported C.37 in late April, it had already been expanding in Chile and Argentina, in the presence of Alpha and Gamma.
virological.org/t/novel-sublin…
17/
virological.org/t/novel-sublin…
17/

C.37 likely originated in Peru in late 2020, given its earlier growth and peak frequency. However, the earliest C.37 record is from Argentina from 8-Nov (EPI_ISL_2158693, although this sequence is identical to the second earliest case, from 29-Jan).
18/
18/
C.37 has accumulated additional mutations of interest appearing multiple times on the global tree: (1) Additional deletion in S:61-75, (2) Q675H, (3) I714V
nextstrain.org/community/quip…
19/


nextstrain.org/community/quip…
19/


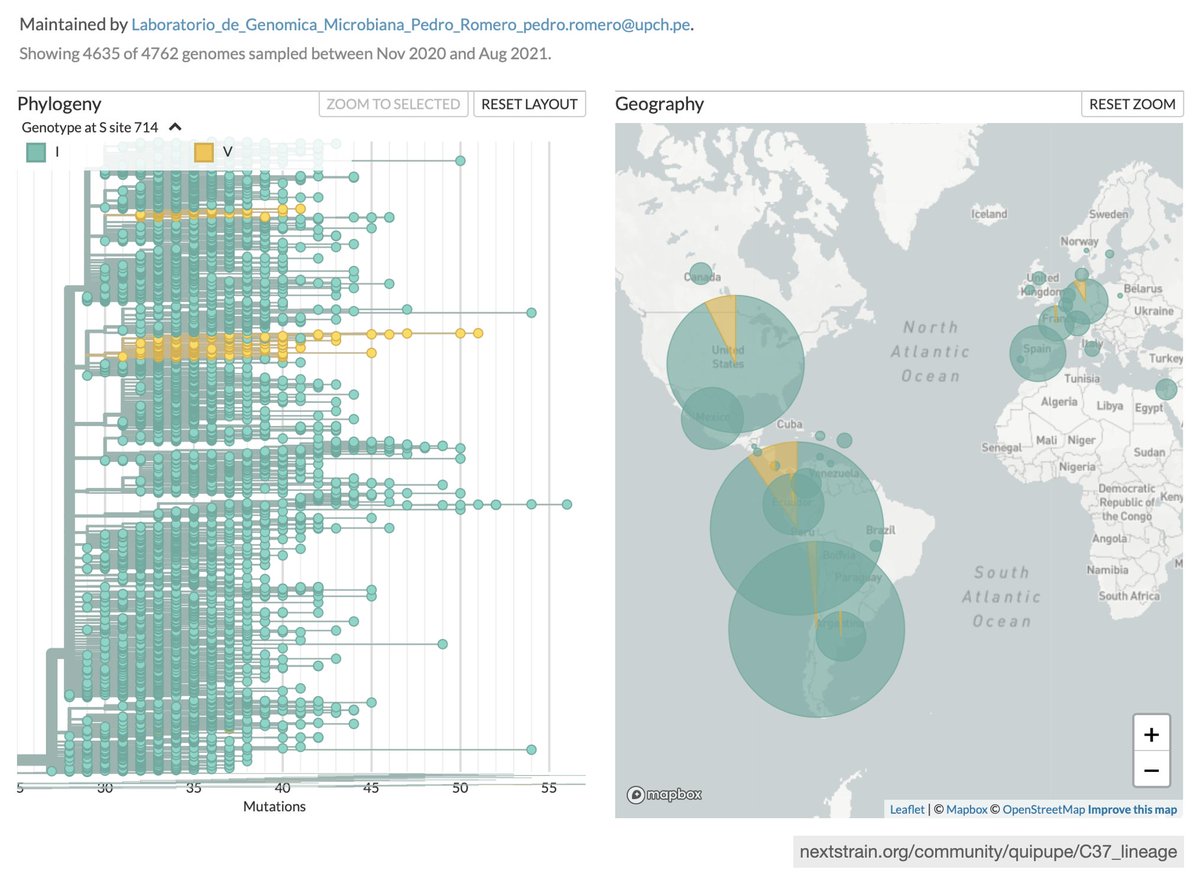
Most countries in Latam have limited capacity for follow-up lab + epi studies, so it's been challenging to assess the effect of C.37 mutations on transmission, virulence, or potential immune scape. Here's a summary of the early evidence from PHE.
assets.publishing.service.gov.uk/government/upl…
20/
assets.publishing.service.gov.uk/government/upl…
20/

Evidence for higher ACE2 affinity + infectivity
biorxiv.org/content/10.110…
medrxiv.org/content/10.110…
21/


biorxiv.org/content/10.110…
medrxiv.org/content/10.110…
21/



Evidence for reduced antibody neutralization to levels similar to Gamma and Delta through Δ246-253, L452Q, F490S:
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
biorxiv.org/content/10.110…
22/



biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
biorxiv.org/content/10.110…
22/




Finally, B.1.621 / Mu. It was first reported in Colombia, with E484K, N501Y, P681H, and a similar emergence pattern and export to other regional variants.
virological.org/t/emergence-of…
medrxiv.org/content/10.110…
23/


virological.org/t/emergence-of…
medrxiv.org/content/10.110…
23/



Colombia has had two epidemic waves in 2021. Gamma + 2020 lineages caused the first peak (Feb), and Mu drove the second peak (June-July).
24/

24/


Data from PHE and a recent preprint suggest reduced neutralization by convalescent + post-vaccine sera to levels greater than Beta, potentially making Mu the most immune-evading variant yet.
biorxiv.org/content/10.110…
assets.publishing.service.gov.uk/government/upl…
25/

biorxiv.org/content/10.110…
assets.publishing.service.gov.uk/government/upl…
25/


Additional evidence on Mu is scarce for similar reasons to Lambda: (1) limited capacity for follow-up studies, and (2) these variants have not been a significant threat in high-income countries like Delta is. The recent VOI designation by WHO should bring new data on Mu soon.
26/
26/
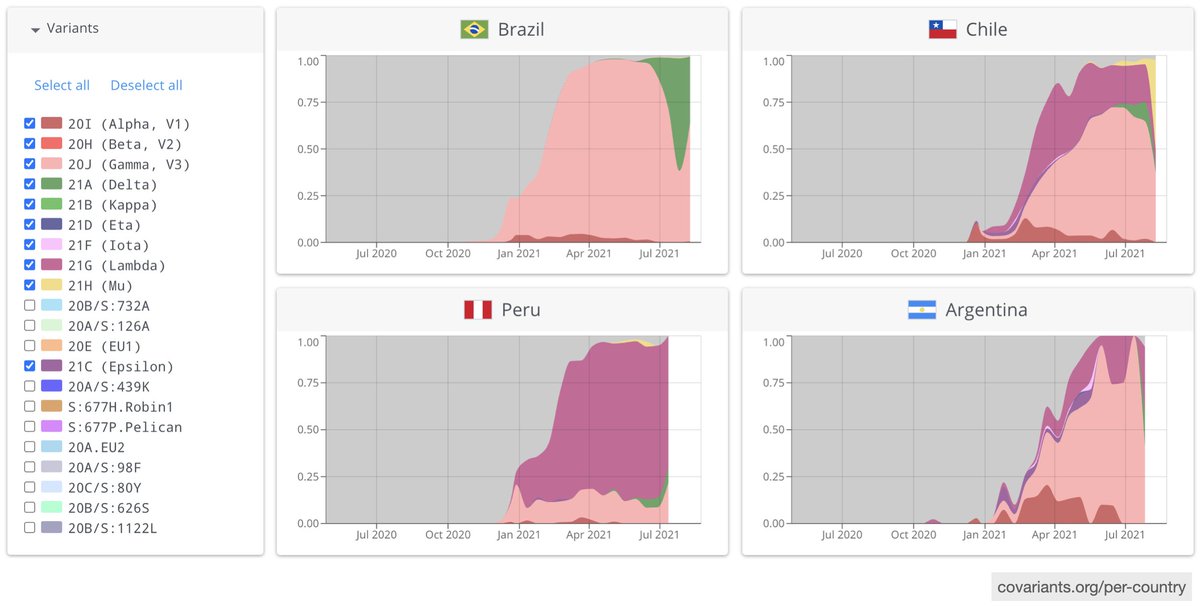
What about other countries? We see diverse mixes of regional variants plus Alpha and other imported lineages in Argentina, Chile, and Ecuador. Other countries report less than 1000 genomes, so it's hard to assess their situation
medrxiv.org/content/10.110…
auspice.cov2.cl/ncov/chile-glo…
27/



medrxiv.org/content/10.110…
auspice.cov2.cl/ncov/chile-glo…
27/




So, it is possible that Gamma, Lambda, Mu, and local variants prevented Alpha from dominating Latam in early 2021, possibly because they could transmit better in populations with high levels of natural immunity from large initial waves of 2020.
28/
28/

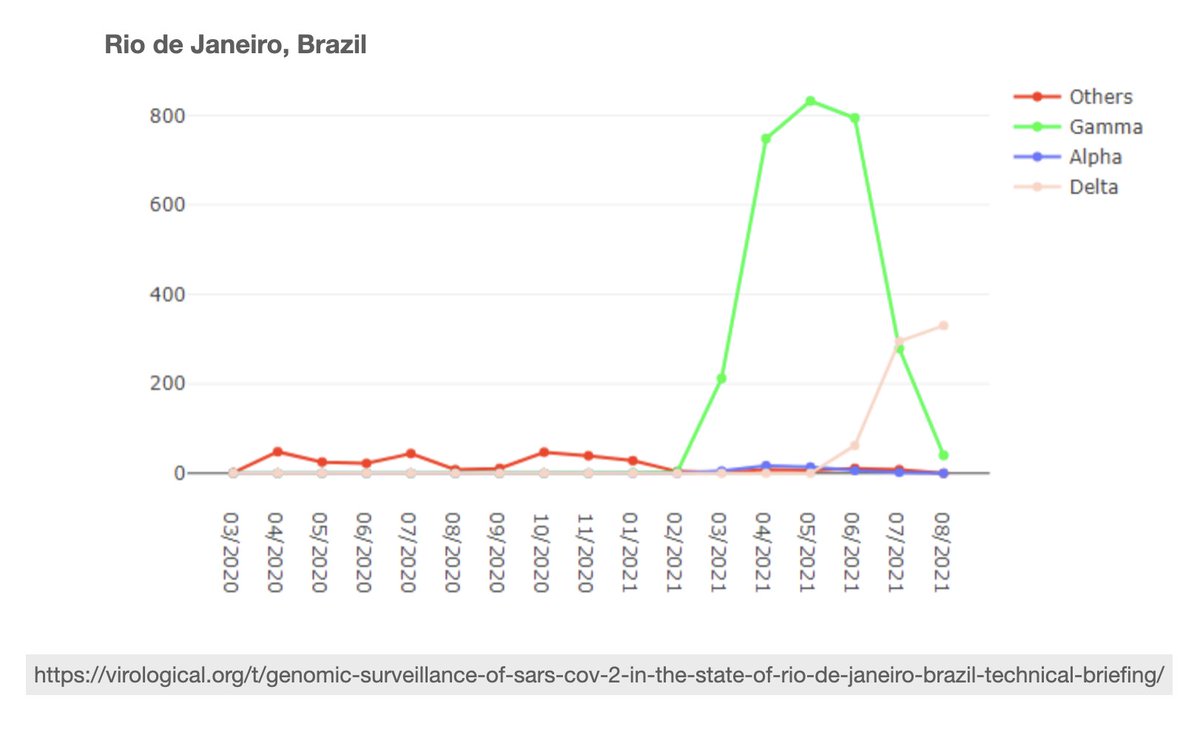
But increased immune evasion may not be enough to overcome Delta's high transmissibility. As new cases fall to their lowest levels of 2021, we see Delta replace Gamma in Rio de Janeiro and Lambda in Lima. Delta is also expanding in Colombia and may replace Mu soon.
29/


29/


Vaccination coverage remains low at ~30%, and most countries are preparing for a rise in cases in the following weeks with the arrival of Delta. So, there's room for novel variants to emerge locally in the next months. We'll see how that goes.
30/
FIN.
30/
FIN.

This is the summary figure I meant to show. From the amazing resource that is CoVariants.org. Gracias, @firefoxx66!
31/
31/

• • •
Missing some Tweet in this thread? You can try to
force a refresh