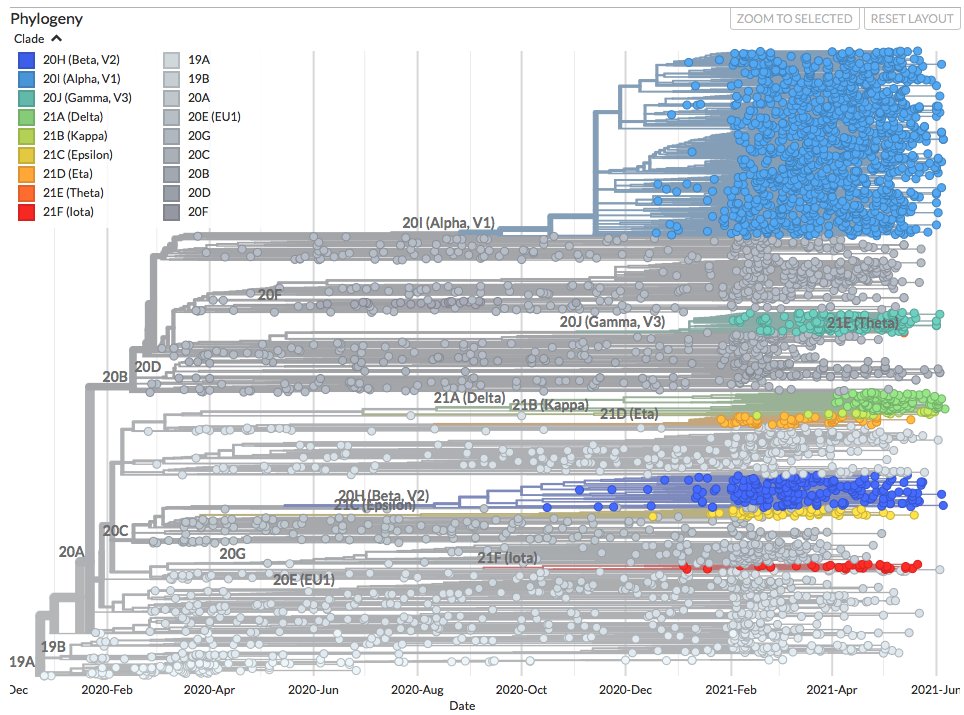
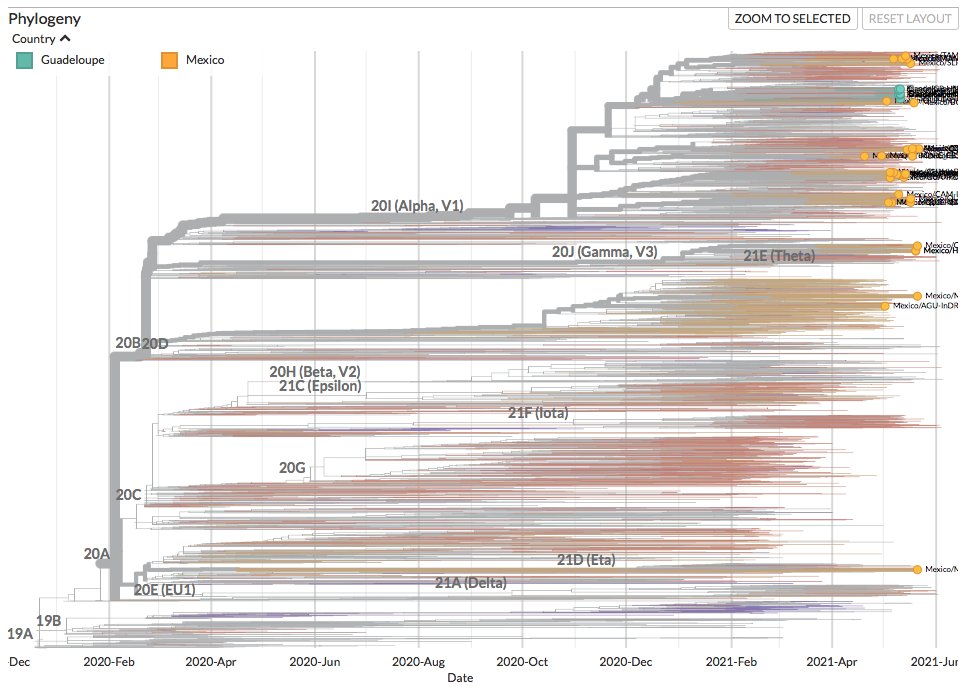
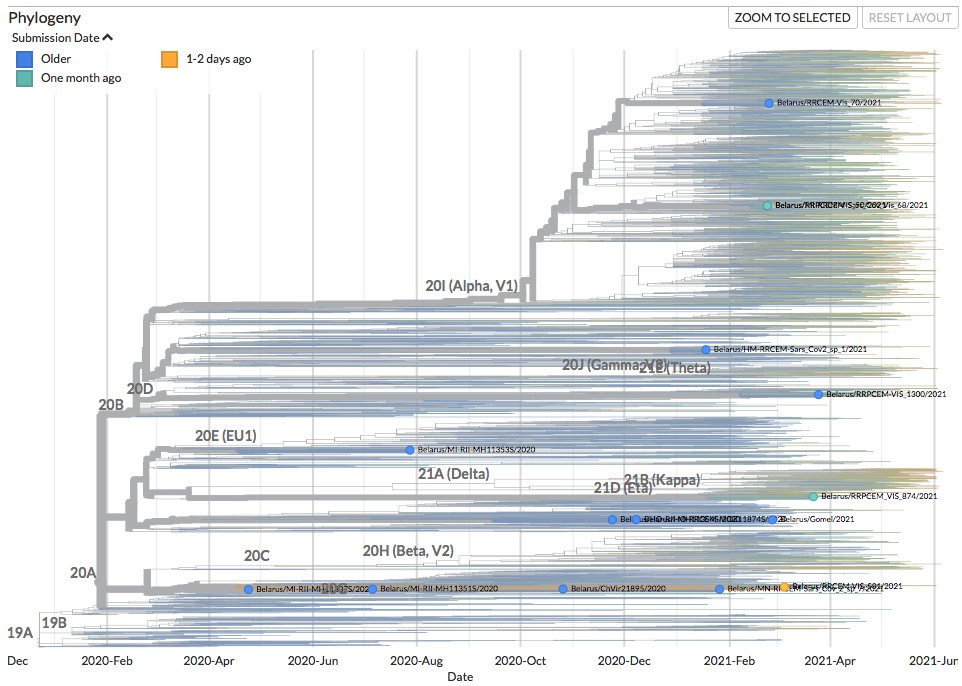
Thanks to #opendata sharing via @GISAID, we've updated nextstrain.org/ncov/gisaid with 741778 new #COVID19 #SARSCoV2 sequences during the last month!
1/7
1/7

Thanks to all new submitters:
NPHL Cameroon, Southwest Hospital Ethiopia, @CerfigGuinee, @NUFeinbergMed, @comui2017, @ILRI, Institut Pasteur Maroc, Central Public Health Reference Lab of Sierra Leone, @PathCareSA1, @CSIR_NEIST, @iitdelhi, @DBEBtweeting
2/7
NPHL Cameroon, Southwest Hospital Ethiopia, @CerfigGuinee, @NUFeinbergMed, @comui2017, @ILRI, Institut Pasteur Maroc, Central Public Health Reference Lab of Sierra Leone, @PathCareSA1, @CSIR_NEIST, @iitdelhi, @DBEBtweeting
2/7
Molecular Solutions Care Health, @MedGenomeLabs, @BjgmcSgh, @IISERPune, @blalbiotech, @uphimpactslives, VenomCoV, @unsri, @DeepSeqNotts, @kmu1928, Nagasaki Institute of Environment and Public Health, Sendai City Institute of Public Health
3/7
3/7
Japan COVID-19 Open Data Consortium, @UTokyo_News_en, Royal Medical Services Jordan, @UKM_UMBI, Medical Research Institute Sri Lanka, @CreativeSoftLK, Kaohsiung Medical University, VG-CARE, Central Research Institute of Epidemiology Belarus
4/7
4/7
Institute of Public Health Bosnia and Herzegovina, Regional Hospital Liberec, University Hospital Pilsen, @UK_Muenster, @UMG_Tweet, Pharmgenetix, @UNI_FIRENZE, @NIPH_RKS, Institute of Public Health of Montenegro, @StichtingPAMM, @WSSEGorzowWlkp, HUGC Dr Negrin
5/7
5/7
@cibir_rioja, @GVAfisabio, @ngs_fisabio, @HGUVALENCIA , SRO Aargau, Synlab Bioggio, @Dynacare, @EasternHealthNL, @UNAM_MX, @SonomaCoHealth, @CUAnschutz, @pucvm, @LifeAtPurdue, @Grambling1901, @Tulane, @wakeforestmed, @UPR_Oficial, @UTHealthSA
6/7
6/7
@UTHealthGCCRI, @UVMLarnerMed, #CICVyA, LACEN PR Brazil, @fmrpusp, MEBLAB, @bioren_ufro, @cienciasuach, @UAustraldeChile, @UdeA, @UCooperativaCol, Gencell Pharma, @CISeAL_PUCE, @UNSA_Oficial
7/7
7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh