What is the relevance of viral load in #COVID19 disease severity? A very talented @YaleMSTP student @SilvaJ_C found that saliva viral load to be a better predictor of disease than nasopharyngeal viral load. Here is a thread to explain the findings. (1/n)
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
I preface by saying that numerous fantastic studies by many have demonstrated nasopharyngeal viral load to correlate with disease severity and mortality, while others do not find such correlation. Here are some of these studies. (2/n) 

Being at @yale, the birthplace of #SalivaDirect and everything saliva bc of awesome colleagues like @awyllie13 @NathanGrubaugh @VogelsChantal et al, we had access to both saliva and nasopharyngeal (NP) samples from the same patients to do direct comparisons. (3/n)
We already knew that saliva is an easy and reliable way to detect viral load in patients, thanks to the great work of colleagues here @YaleSPH @YaleMed. @awyllie13 et al. (4/n)
nejm.org/doi/full/10.10…
nejm.org/doi/full/10.10…
What about saliva’s ability to predict #COVID19 disease? @SlivaJ_C compared first whether saliva and NP viral load correlates with risk factors, age, sex. Saliva (top panels) but not NP (btm panels) viral load correlated with all of these. (5/n) 

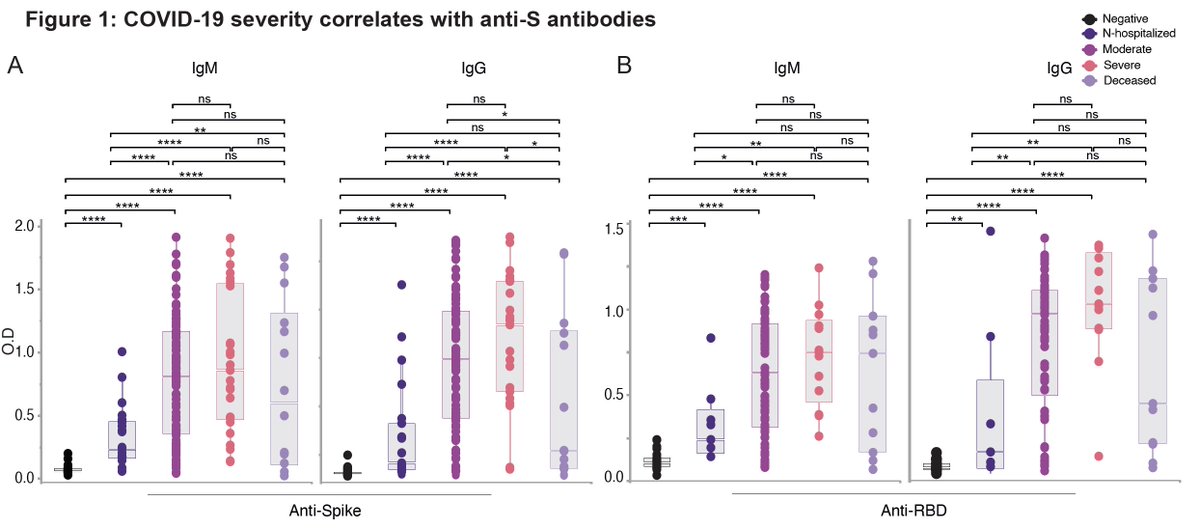
Next, @silvaJ_C examined whether viral loads correlate with severity of #COVID19. Saliva (top panels) but not NP (btm panels) viral load was elevated in hospitalized, severe and deceased patients. (6/n) 

Moreover, saliva viral load (top) and NP viral load (btm) declined over time in patients that survived #COVID19 but not those who died (purple). Patients with lethal disease maintained v. high viral load over the course of disease. (7/n) 

What associates with saliva viral load? Most influential predictors of saliva viral load was assessed by running a non-linear iterative
partial least squares (NIPALS) analysis of all factors and then assessing for variable importance with Variable Importance Plot (VIP). (8/n)
partial least squares (NIPALS) analysis of all factors and then assessing for variable importance with Variable Importance Plot (VIP). (8/n)

Positive correlation with saliva load: the usual suspects-chemokine, cytokines, IFNs.
Negative correlation with saliva load: antiviral antibodies and platelets.
We know early antibody kinetics is key to survival @carolilucas @sneakyvirus1 et al. (9/n)
medrxiv.org/content/10.110…
Negative correlation with saliva load: antiviral antibodies and platelets.
We know early antibody kinetics is key to survival @carolilucas @sneakyvirus1 et al. (9/n)
medrxiv.org/content/10.110…
Patients with high saliva viral load developed antiviral antibodies with significantly delayed kinetics than those who had low viral load. (10/n) 

Time spent in the high, medium, and low saliva viral
load brackets was compared. Deceased
patients spent longest time in the high saliva viral load bracket, while non-hospitalized spent shortest time with high viral load. Moderate/severe in between. (11/n)
load brackets was compared. Deceased
patients spent longest time in the high saliva viral load bracket, while non-hospitalized spent shortest time with high viral load. Moderate/severe in between. (11/n)

Why is saliva viral load a better correlate of disease than NP load? We think that NP only reflects upper respiratory tract (URT) viral replication, while saliva represents both URT and lower respiratory tract. Mucociliary clearance propels virus from LRT to oral cavity. (12/n) 

URT virus is key to transmission, but LRT is key to severe #COVID19. Saliva appears to be better at capturing the critical viral source driving severe disease.
This was once again an amazing collaboration with the Yale IMPACT team 🙏🏼 Led by @SilvaJ_C 💪🏼 (end)
This was once again an amazing collaboration with the Yale IMPACT team 🙏🏼 Led by @SilvaJ_C 💪🏼 (end)
• • •
Missing some Tweet in this thread? You can try to
force a refresh