
The Road to Paxlovid (PF-07321332)
A thread. #paxlovid #protease #inhibitors #ivermectin #COVID19
Dr. John Campbell: Paxlovid vs Ivermectin
A thread. #paxlovid #protease #inhibitors #ivermectin #COVID19
Dr. John Campbell: Paxlovid vs Ivermectin
New Pfizer antiviral and ivermectin, a pharmacodynamic analysis. Paxlovid (PF-07321332), C₂₃H₃₂F₃N₅O₄
Paxlovid is designed to block the activity of the SARS-CoV-2-3CL protease, an enzyme that the coronavirus needs to replicate. Co-administration with a low dose of
Paxlovid is designed to block the activity of the SARS-CoV-2-3CL protease, an enzyme that the coronavirus needs to replicate. Co-administration with a low dose of
ritonavir helps slow the metabolism, or breakdown, of Paxlovid in order for it to remain active in the body for longer periods of time at higher concentrations to help combat the virus. Paxlovid inhibits viral replication at a stage known as proteolysis, which occurs before
viral RNA replication. In preclinical studies, Paxlovid did not demonstrate evidence of mutagenic DNA interactions.
Ritonavir can boost other drug by inhibiting Cytochrome p450.
pfizer.com/news/press-rel…
Ritonavir can boost other drug by inhibiting Cytochrome p450.
pfizer.com/news/press-rel…
This study employed an enzymatic assay for qHTS that identified 23 SARS-CoV-2 3CLpro inhibitors from a collection of approved drugs, drug candidates, and bioactive compounds. These 3CLpro inhibitors can be combined with drugs of different targets to evaluate their potential in
drug cocktails for the treatment of COVID-19. In addition, they can also serve as starting points for medicinal chemistry optimization to improve potency and drug-like properties. pubs.acs.org/doi/pdf/10.102…
The activity of the anti-SARS-CoV-2 viral infection was confirmed in 7 of 23 compounds. Microscopic interactions between Ivermectin and key human and viral proteins involved in SARS-CoV-2 infection. By using blind docking and extensive MD simulations, at a molecular and
atomic resolution, the physicochemical interactions of ivermectin with key human and viral protein targets active in the SARS-CoV-2 infection cycle. The flexibility induced by the fused rings of the drug, in combination with the multiple polar groups, such as alcohols and
esters, allow efficient non-covalent interactions with some parts of the protein targets, especially of hydrogen bonding type. Ivermectin to be most suitable in blocking the ACE2/RBD complex formation, bind with the CLPro and PLPro. pubs.rsc.org/en/content/art…
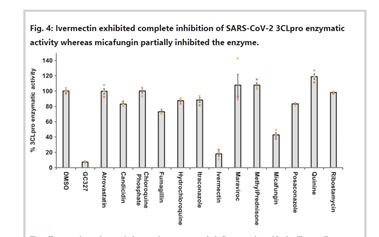
Identification of 3-chymotrypsin like protease (3CLPro) inhibitors as potential anti-SARS-CoV-2 agents. The strength and persistency of the interaction between Ivermectin and the binding site of 3CLpro indicate that a partial inhibition of the catalytic activity could have place
as the drug interacts with the main subdomains that define the enzyme binding pocket. Identification: boceprevir, micafungin, ombitasvir, paritaprevir, and tipranavir exhibited partial inhibitory effect,
whereas Ivermectin was able to completely inhibit the SARS-COV-2 3CLpro enzymatic activity in vitro at the tested doses. 


Ivermectin interacts with the spike glycoprotein of SARS-CoV-2. It has an intense binding of both Ivermectin B1a and B1b isomer to the MPro, interacts with viral replicase (Nsp9), human ACE2 receptor protein, human TMPRSS2 receptor protein. 

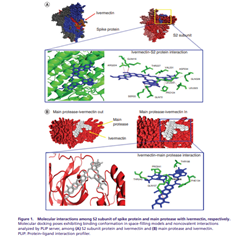

Ivermectin efficiently binds to the viral S protein as well as the human cell surface receptors ACE-2 and TMPRSS2; therefore, it might be involved in inhibiting the entry of the virus into the host cell. It also binds to Mpro and PLpro of SARS-CoV-2; therefore,
it might play a role in preventing the post-translational processing of viral polyproteins. The highly efficient binding of ivermectin to the viral N phosphoprotein and nsp14 is suggestive of its role in inhibiting viral replication and assembly. frontiersin.org/articles/10.33…
With SARS-CoV-2 S Spike protein Ivermectin showed high binding affinity to the viral S protein as well as the human cell surface receptors ACE-2 and TMPRSS2. In agreement to our findings, ivermectin was found to be docked between the viral spike and the ACE2 receptor Binding
Interactions of Selected Drugs With Human TMPRSS2 Protein (ACE2 protein). The docking results revealed that ivermectin showed the highest binding affinity to the active site of the protein (MolDock score −174.971) and protein–ligand interactions . Binding Interactions of
Selected Drugs With Human ACE-2 Protein that ivermectin showed the highest binding affinity to the active site of the protein (MolDock score −159.754) and protein–ligand interactions. With SARS-CoV-2 S Glycoprotein Ivermectin showed the highest binding affinity to the predicted
active site of the protein. With SARS-CoV-2 Nsp14 Protein ivermectin showed the highest binding affinity (MolDock score −212.265) and protein–ligand interactions. Binding Interactions of Selected Drugs With SARS-CoV-2 PLpro.
Ivermectin showed the highest binding affinity to the predicted active site of the protein (MolDock score −180.765) and protein–ligand interactions.
• • •
Missing some Tweet in this thread? You can try to
force a refresh











