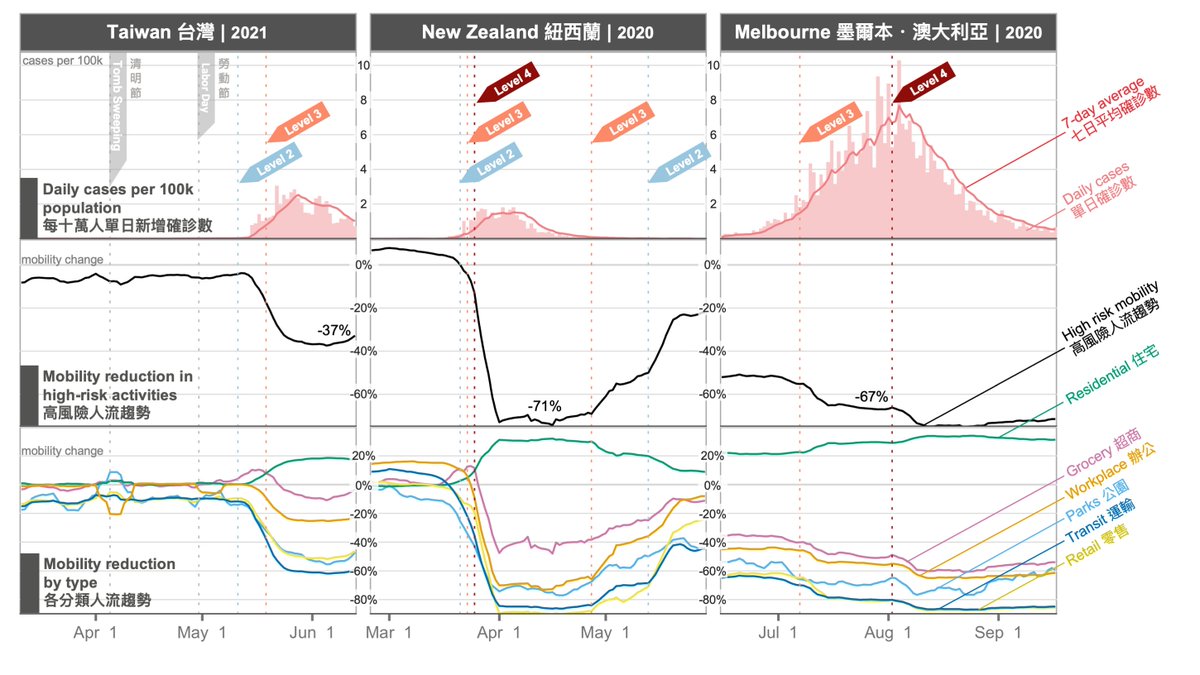
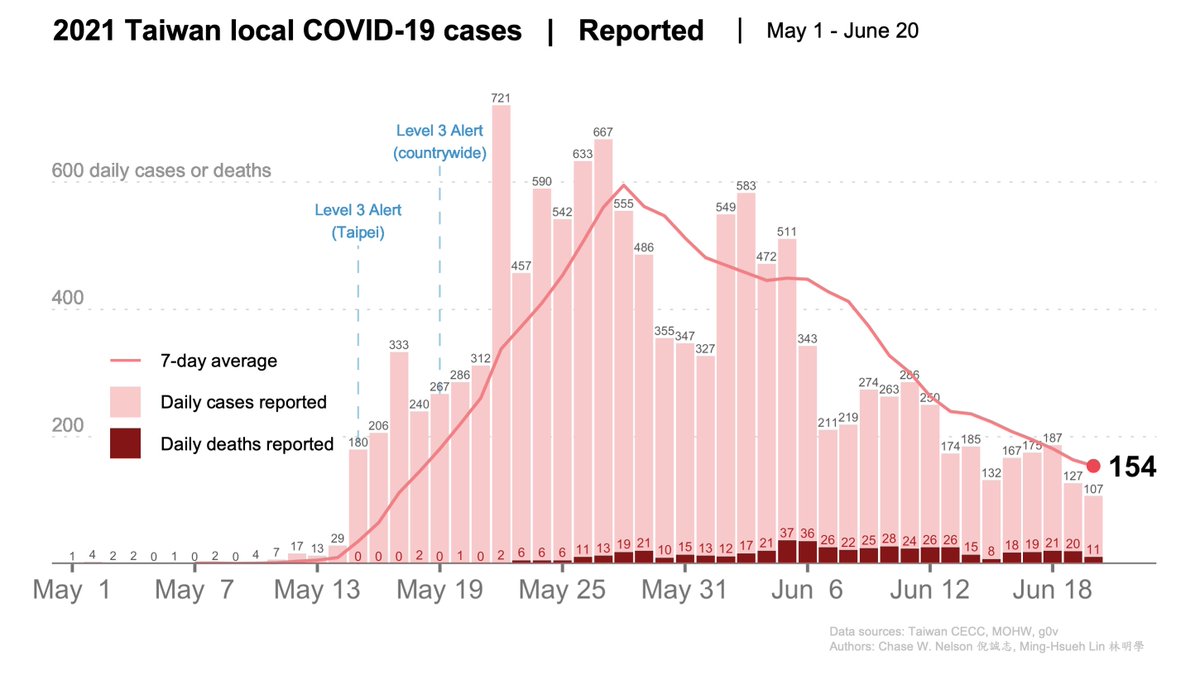
TODAY at #Virological: throughout the #COVID19 pandemic, there has been a tendency to underestimate the potential of #SARSCoV2 to mutate and adapt.
As the @US_FDA meets Tuesday to discuss #molnupiravir, @sarperotto and I fear another oversight.🧵
1/15
virological.org/t/mutagenic-an…
As the @US_FDA meets Tuesday to discuss #molnupiravir, @sarperotto and I fear another oversight.🧵
1/15
virological.org/t/mutagenic-an…
To be clear, I believe vaccines and antiviral drugs BOTH have critical parts to play in the fight against #COVID19.
Oral antivirals are of particular interest given their potential for equitable distribution.
This is NOT an argument against antivirals.
2/15
Oral antivirals are of particular interest given their potential for equitable distribution.
This is NOT an argument against antivirals.
2/15
Antivirals can work via several mechanisms.
🦠PROTEASE INHIBITORS, like #paxlovid and #masitinib, prevent the production of mature viral proteins.
🧬MUTAGENS, like #molnupiravir, instead increase the viral mutation rate to intolerable levels, causing LETHAL MUTAGENESIS.
3/15
🦠PROTEASE INHIBITORS, like #paxlovid and #masitinib, prevent the production of mature viral proteins.
🧬MUTAGENS, like #molnupiravir, instead increase the viral mutation rate to intolerable levels, causing LETHAL MUTAGENESIS.
3/15
Critically, any self-administered drug risks low concentrations due to missed doses, incomplete courses, or low initial penetrance.
For #molnupiravir, this might induce only SUBLETHAL MUTAGENESIS, accelerating within-host virus evolution and potentiating new variants.
4/15
For #molnupiravir, this might induce only SUBLETHAL MUTAGENESIS, accelerating within-host virus evolution and potentiating new variants.
4/15
The risk of mutagens is underscored by the fact that another antiviral, #ribavirin, induces adaptive mutations in other RNA viruses. Moreover, past #SARSCoV2 variants of concern likely acquired adaptive combinations of mutations during chronic infections before transmitting.
5/15
5/15
Given the potential for SUBLETHAL MUTAGENESIS of #SARSCoV2 by #molnupiravir, steps should be taken to understand the evolutionary consequences of low drug concentrations and improper administration for pathogen evolution.
Below is a list of issues to consider.👇
6/15
Below is a list of issues to consider.👇
6/15
1⃣ Because #molnupiravir will be taken by patients with recent symptom onset, and drug concentration increases from 0 over time, PEAK VIRAL SHEDDING is likely to occur while drug concentration is still low. This could potentiate the shedding of mutant — but viable — virus.
7/15
7/15
2⃣ #Molnupiravir has a SHORT PLASMA HALF-LIFE, making low concentrations easier to achieve, say, as a result of missed or inconsistently timed doses.
8/15
8/15
3⃣ Coronaviruses have a propensity for RECOMBINATION, which can help purge viral genomes of deleterious mutations, or generate adaptive combinations of beneficial or compensatory mutations. These phenomena are therefore critical to include in models and simulations.
9/15
9/15
4⃣ #SARSCoV2 has a PRE-EXISTING BIAS for C→U mutations — the main mutation caused by #molnupiravir — as well as a genomic G:C content of 38%, and a plus-strand C content of 18%. These characteristics limit the extent to which molnupiravir can raise the mutation rate.
10/15
10/15
5⃣ Epidemiological spread of adaptive mutations is limited by both:
(a) their GENERATION VIA MUTATION within hosts; and
(b) the TRANSMISSION BOTTLENECK SIZE between hosts, that is, the number of viral genomes that found a new infection.
11/15
(a) their GENERATION VIA MUTATION within hosts; and
(b) the TRANSMISSION BOTTLENECK SIZE between hosts, that is, the number of viral genomes that found a new infection.
11/15
6⃣ Even if the average number of mutations per viral genome is high, the DISTRIBUTION OF MUTATION COUNTS can still include a class of minimally mutated viable genomes. This could be due to the mutation mechanism or ‘compartments’ with low drug concentrations within a host.
12/15
12/15
7⃣ The MUTATIONAL ROBUSTNESS of #SARSCoV2 is not known. The highest mutation rate tolerated by a virus, just possibly 1-5 per replication per genome, depends on the size of the functional genome and the expendability of ‘accessory’ genes.
13/15
13/15
EVERYONE must understand the importance of taking #molnupiravir as directed, and of quarantining while doing so, as the above issues are studied.
We must ensure that #SARSCoV2 is not inadvertently handed mutational resources for the accelerated generation of new variants.
14/15
We must ensure that #SARSCoV2 is not inadvertently handed mutational resources for the accelerated generation of new variants.
14/15

🙏THANKS to coauthor @sarperotto and all who generously gave feedback: @zachary_ardern, @AprilWei001, @GoldbergTony, @jbloom_lab, and @RJABuggs, who first raised this issue to me on October 6 and provided invaluable insight and edits.
All views and any errors are our own.
15/15
All views and any errors are our own.
15/15
@sarperotto @zachary_ardern @AprilWei001 @GoldbergTony @jbloom_lab @RJABuggs 🚨UPDATE on #MOLNUPIRAVIR.
Below is a review of the @US_FDA materials and November 30 panel discussion on the potential for this drug to cause new #SARSCoV2 variants of concern.
Overall, the panel voted in favor of authorization, 13 YES✅ to 10 NO❌.
👇
Below is a review of the @US_FDA materials and November 30 panel discussion on the potential for this drug to cause new #SARSCoV2 variants of concern.
Overall, the panel voted in favor of authorization, 13 YES✅ to 10 NO❌.
👇
https://twitter.com/chasewnelson/status/1466331830345293827
Some conceptual figures illustrating our concerns are here👇
https://twitter.com/chasewnelson/status/1466722854876377093
Additional insights from @LauringLab and others on whether the mutation-inducing antiviral #molnupiravir could accelerate #SARSCoV2 variants of concern👇
https://twitter.com/chasewnelson/status/1468164091814748161
🦠💊UPDATE on #Molnupiravir: paper out in @NEJM.
One sentence addresses the concern that its poorly characterized mutagenicity could accelerate virus evolution: "The mechanism of action of molnupiravir is independent of mutations in the spike protein..."

One sentence addresses the concern that its poorly characterized mutagenicity could accelerate virus evolution: "The mechanism of action of molnupiravir is independent of mutations in the spike protein..."
https://twitter.com/NEJM/status/1471604809396482048

It is correct that the drug should work the same regardless of Spike genotype.
A false interpretation is that molnupiravir doesn't mutate Spike. It does, along with the rest of the genome.
Thus, I believe we still have only two relevant data summaries from Merck:
A false interpretation is that molnupiravir doesn't mutate Spike. It does, along with the rest of the genome.
Thus, I believe we still have only two relevant data summaries from Merck:
1⃣ Merck reports a mean of ~7.5 mutations at a within-patient frequency of >5% across the virus genome.
Note the range includes *0*. Note too this is within-host diversity (polymorphism) filtered by selection, not a mutation rate.
Note the range includes *0*. Note too this is within-host diversity (polymorphism) filtered by selection, not a mutation rate.
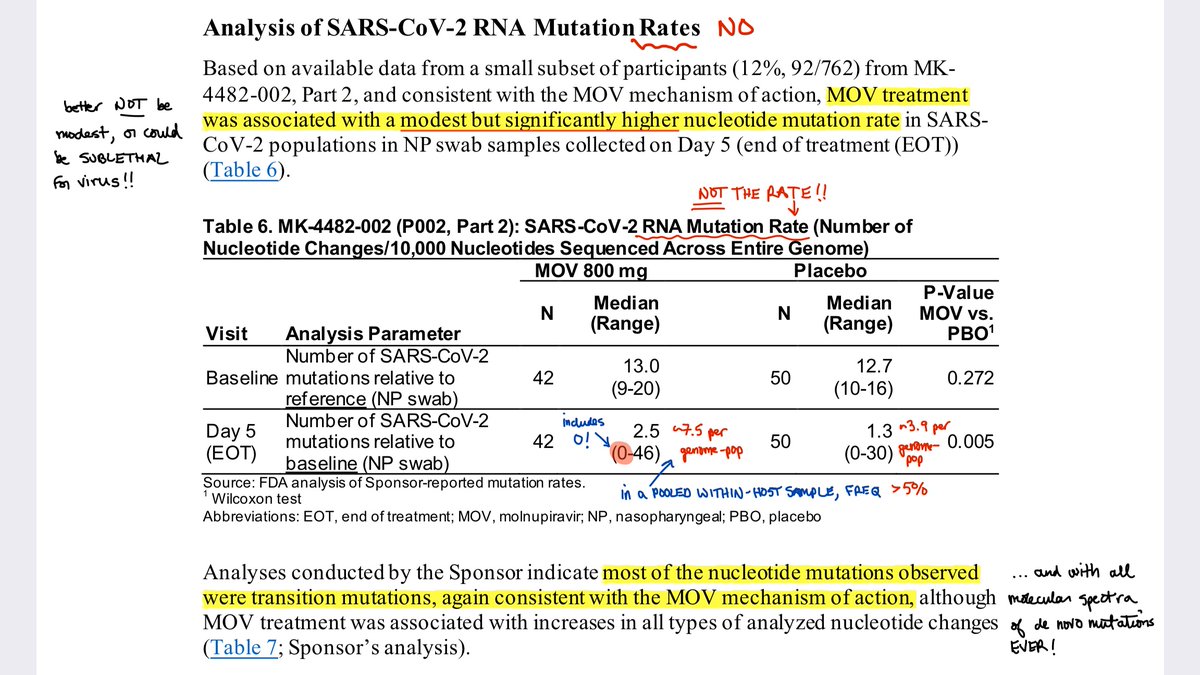
2⃣ Merck provides this chart, also difficult to interpret, but again likely referring to within-host variants at >5% frequency. Note that Baseline and Day 5 distributions *overlap*.
More is needed, including raw sequence data, to estimate virus mutation and onward transmission.
More is needed, including raw sequence data, to estimate virus mutation and onward transmission.

• • •
Missing some Tweet in this thread? You can try to
force a refresh