
Really chuffed to share our latest work from Sanjana Lab out in @nature today (nature.com/articles/s4158…).
We tested >12,000 genes to find positive regulators of T cell proliferation to be used for next-gen #immunotherapies.
A thread...
We tested >12,000 genes to find positive regulators of T cell proliferation to be used for next-gen #immunotherapies.
A thread...
(2/28) T cell therapies have shown the potential to cure patients from #cancer – but even in B cell cancers, relapses outnumber cures… One of the problems is limited T cell persistence. #celltherapy #CARTcell
(3/28) We wanted to find new genes (including genes never expressed in T cells) that could improve T cell persistence. Now, the question was: should we use #crispr activation or directly deliver target genes on a #lentivirus?
(4/28) As always, months of optimisation followed. Eventually, lentiviral overexpression proved to be more robust than CRISPRa in our hands. And thanks to @teresadavoli3, we had access to a DNA-barcoded lentiviral library containing over 12,000 full length human genes.
(5/28) We delivered the barcoded gene library to CD4+ and CD8+ T cells from several healthy donors and sorted the cells that proliferated best in response to stimulation. 

(6/28) We quantified the gene-linked barcode abundance in most proliferating T cells and compared it to the unselected population. Seeing independent enrichment of multiple barcodes for the same gene gave us confidence in our selection of top-ranked genes. 

(7/28) We validated the top-ranked genes from our screen in new independent donors. In fact, we found out that the genes which improved proliferation also improved other T cell functions (which was also an opportunity to include a #rainbow plot in the paper #lgbtqstem). 
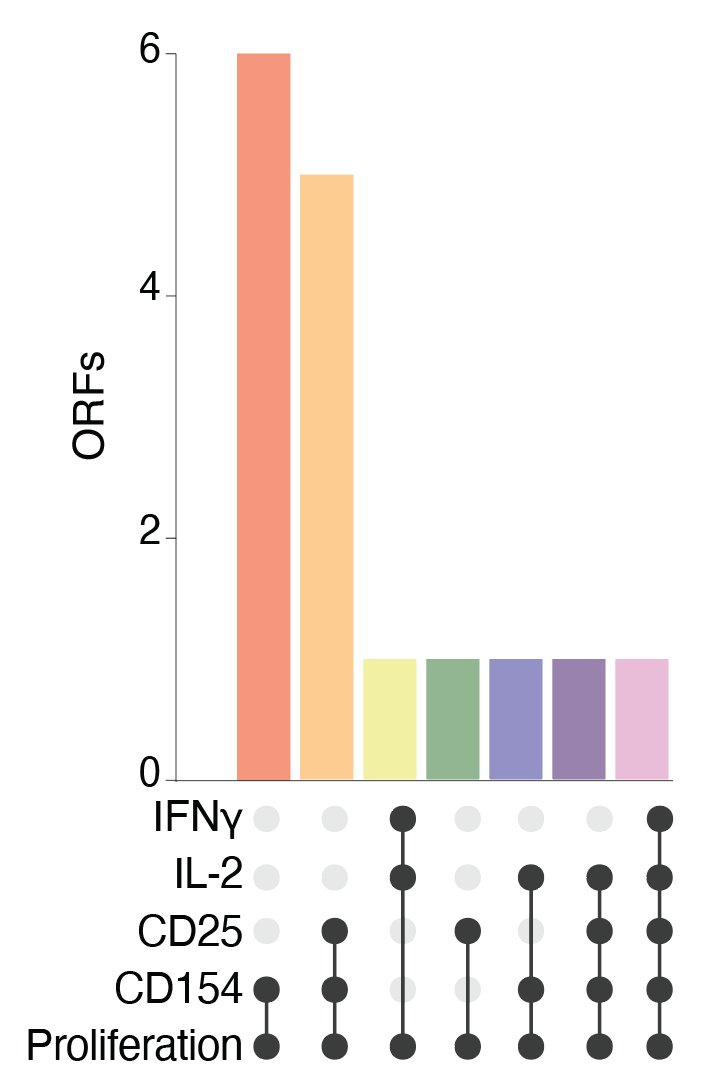
(8/28) What was particularly interesting, the top-ranked screen gene LTBR drastically increased secretion of two key T cell #cytokines, interferon gamma and interleukin 2. 

(9/28) Taking advantage of our next-door neighbours @nygctech and their experience in #singlecell technologies, we developed a #multiomics method to run an overexpression screen in T cells with a single cell transcriptome and surface antigen readout – OverCITE-seq. #scRNAseq 

(10/28) With OverCITE-seq, we were able to map transcriptomic changes induced by ~30 overexpressed genes in resting and stimulated T cells. Key finding: LTBR overexpression had a really powerful effect on T cell state. 

(11/28) So what is LTBR and how does it work in T cells?
(12/28) According to literature, LTBR, or lymphotoxin beta receptor, plays a key role in formation of secondary lymphoid organs (for instance lymph nodes) and can be expressed by diverse cell types – but not T cells or other lymphocytes.
(13/28) We first looked at #transcriptomes of T cells transduced with LTBR and compared them to matched control. LTBR overexpression let to upregulation of hundreds of genes, many of them involved in important T cell processes. 

(14/28) We validated many of the processes identified on the #transcriptome level with functional assays and found out that LTBR improved several aspects of T cell phenotype and function (which we did not screen for!).
(15/28) For instance, LTBR protected T cells from apoptosis and dysfunction after chronic stimulation. LTBR T cells also retained a younger, less differentiated phenotype and expressed high levels of TCF1, a key transcription factor of T memory stem cells. #Tcellexhaustion 

(16/28) Given the profound transcriptional changes observed in LTBR T cells, we wanted to see if they were accompanied by #epigenetic rewiring. We found out that in LTBR T cells #chromatin was more open at regions strongly enriched for the NFkB binding motifs. 

(17/28) Then, we examined the (phospho)protein levels of key mediators in the LTBR/NFkB pathway and found an increased activation of both the canonical and non-canonical NFkB. #SignalingPathways #NFKB 

(18/28) More questions remained. Were both canonical and non-canonical NFkB pathways important for the LTBR phenotype, or was one more important than the other? To address that, we developed a combinatorial gain-of-function and loss-of-function assay. #crispr #NFKB 

(19/28) By co-delivering LTBR or a control gene, and guide RNAs targeting the key mediators of canonical and non-canonical NFkB pathways we showed that only the canonical NFkB drove the transcriptional changes in LTBR T cells. #crispr #NFKB 

(20/28) Finally, we combined our top-ranked genes with two #fda approved #CART constructs: #tisacel (4-1BB costim) and #axicel (CD28 costim), to test if we could increase antigen-specific reactivity. 

(21/28) For 6/7 genes tested, we observed increased IFNg and IL2 secretion in response to CD19+ #Bcell #leukaemia cells and enhanced #cytotoxicity. See video below: cancer cells are labelled in green, “enhanced” CAR T cells co-express LTBR.
(22/28) All the work so far was done in T cells from healthy donors. But what about T cells from cancer patients, typically more dysfunctional?
(23/28) We transduced our CARs, with or without LTBR, into T cells from diffuse large B cell lymphoma patients – and observed an increase of effector functions, even in context of #autologous CD19+ target cells. #DLBCL #CART 

(24/28) Out of curiosity, we decided to test if top-ranked genes from a screen in conventional ab T cells would translate to unconventional gd T cells - which recognise conserved stress-induced antigens on diverse types of cancer without MHC restriction. #gdTcell
(25/28) Surprisingly, genes identified in an ab T cell screen improved antitumour response of gd T cells – in context of blood and solid tumours. #gdTcell #immunotherapy #PDAC #PancreaticCancer 

Image by @Ella_Maru
(26/28) #TLDR: a gain of function screen for drivers of proliferation in primary human T cells allowed us to discover new regulators of T cell functions that may potentially be used to improve new and existing #immunotherapies. 

(27/28) This study was only possible thanks to my fantastic colleagues and collaborators Zoran Gajic, Maria Guarino, @ZDaniloski, @jahanpehechanho, @XueXinhe, @congyilv, @_llu_0112, @emimitou, @slhao, @TeresaDavoli3, Catherine Diefenbach, @psmibert, and @nevillesanjana. 

(28/28) Last but not least – I want to thank our institutions (@nygenome @nyuniversity @nyulangone@Perlmutter_CC) and funders @NIH @CancerResearch, and @HopeFunds1 for funding my #postdoc #fellowship.
Photo by @vickyle
• • •
Missing some Tweet in this thread? You can try to
force a refresh



