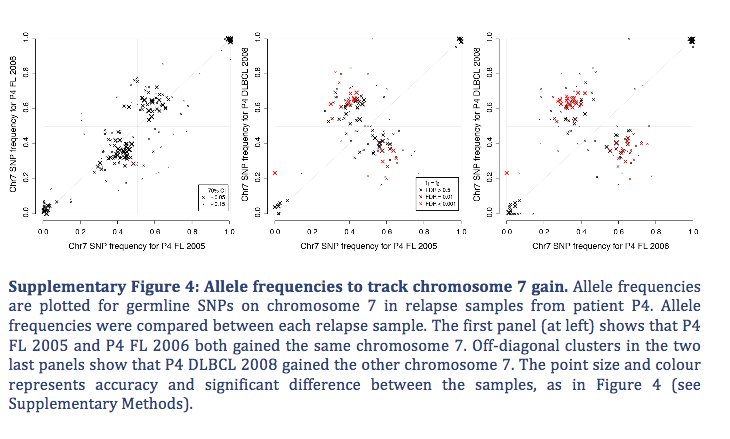
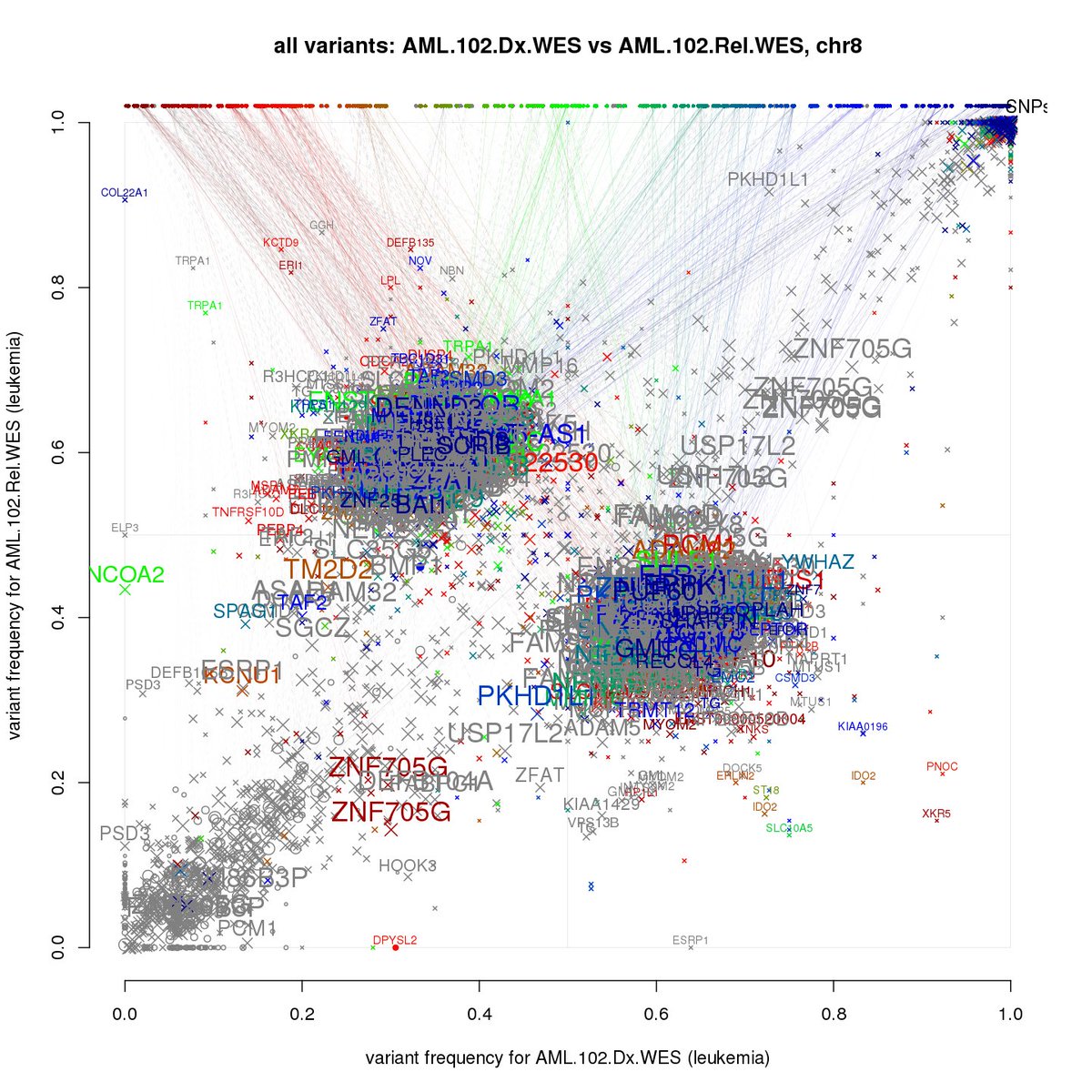
If you want to use: github.com/ChristofferFle…
I've developed and used this the last 4+ years, and now finally out! 🎉
Let me make a thread (or tree?) from here.
It analyses cancer exomes, especially geared for multiple matched cancer samples.
- calls somatic SNVs
- calls CNAs
- calls clones and phylogeny
as well as quality of life:
- annotates variants (effect, dbSNP, ExAC, COSMIC)
- lots of plots: results and QC
Not because it beats some other method in precision/recall on simulated data, but because it's been developed to answer questions from ours and others data. So focus has been on:
- robust to noise
- working without matched normal
- lots of QC