"Very rare pathogenic genetic variants detected by SNP-chips are usually false positives: implications for direct-to-consumer genetic testing"
biorxiv.org/content/10.110…
Let's start...
👇👇👇
So what's the issue?
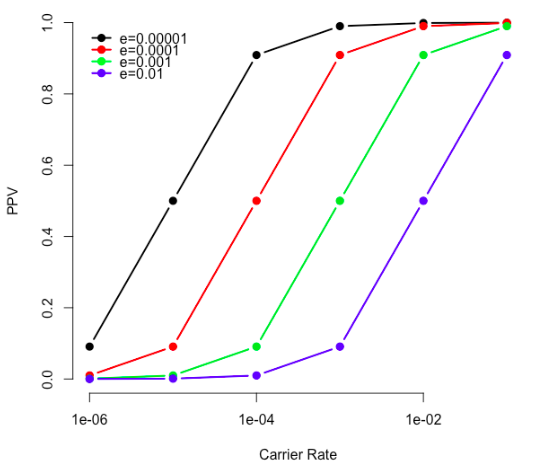
So let's summarise, we validate every positive using Sanger, our array combines multiple probes, we do not test SNPs that are likely to be erroneous, and we use a different array technology.