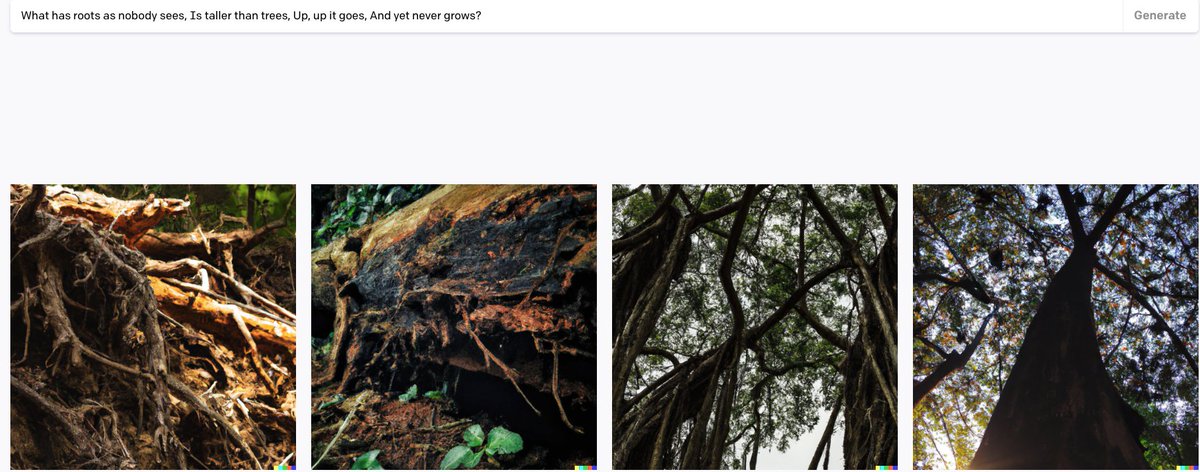
Can DALLE answer riddles from the Hobbit? Let's find out!
#dalle #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI
#dalle #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI
(Except that the answer is mountains, not trees)
Honestly, butterflies and insects isn't bad. But it's hard to draw "Wind" #dalle2 #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddles 

It got dark right!
(Some of the riddles were filtered out by censored keywords - kill, ends life, slay)
#dalle #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #dark #fantasy #hobbit #riddles
(Some of the riddles were filtered out by censored keywords - kill, ends life, slay)
#dalle #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #dark #fantasy #hobbit #riddles

Water is indeed #cold and #clammy, but it's not #fish!
#dalle #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddles #hobbit #tolkien #VQA
#dalle #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddles #hobbit #tolkien #VQA

Would you count the first image as #time?
#dalle2 #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddles #hobbit #tolkien #VQA
#dalle2 #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddles #hobbit #tolkien #VQA

Prompt engineering did not improve things #prompt #promptengineering
#dalle2 #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddle #computerart
#dalle2 #openai #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #riddle #computerart

Bonus: I tried using to make me a shareable image for our protein language model work. Didn't work 😛 (And text is hard for it!).
doi.org/10.1093/bioinf…
#dalle #openai #experiment #dalle2 #aiart #figure #protein #bioinformatics #proteinBERT #bert #languagemodel #science
doi.org/10.1093/bioinf…
#dalle #openai #experiment #dalle2 #aiart #figure #protein #bioinformatics #proteinBERT #bert #languagemodel #science

What have I got in my pocket?
#dalle #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #ring #MachineLearning #ddofer
#dalle #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #ring #MachineLearning #ddofer

"What has the Hobbit got in it's pocket?"
#dalle #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #ring #MachineLearning #ddofer #pocket #art
#dalle #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #ring #MachineLearning #ddofer #pocket #art

"Draw what the Hobbit has in its pocket" #dalle2 #openai #experiment #dalle2 #aiart #AiArtwork #hobbit #LOTR #AI #ring #MachineLearning #ddofer #pocket #art #Hobbit 

Bonus: From @OpenAI 's #DALLE : "An epic fantasy drawing of Mount Doom, in Morgoth from LOTR, in the style of Thirty-six Views of Mount Fuji (Hiroshige)"
#dalle2 #aiart #mordor #fuji #art #dalle #tolkien #LOTR
#dalle2 #aiart #mordor #fuji #art #dalle #tolkien #LOTR

• • •
Missing some Tweet in this thread? You can try to
force a refresh