Let's use this thread to talk about #DNAdatastorage in general and finish up with our work and some thoughts about the future of the field.
1/10
As the burden of the #BigData era on data storage infrastructure is becoming too heavy we need to explore alternatives.
DNA was optimized as an information carrier by billions of years of evolution. It's dense, stable, and energetically efficient.
2/10
Since, others has shown encoding/technological improvements
3/10
4/10
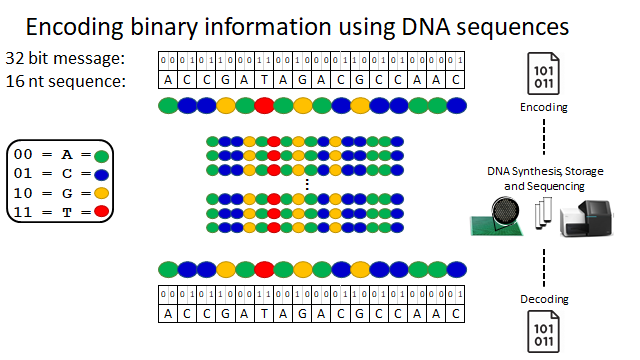
This is where our work comes in.
5/10
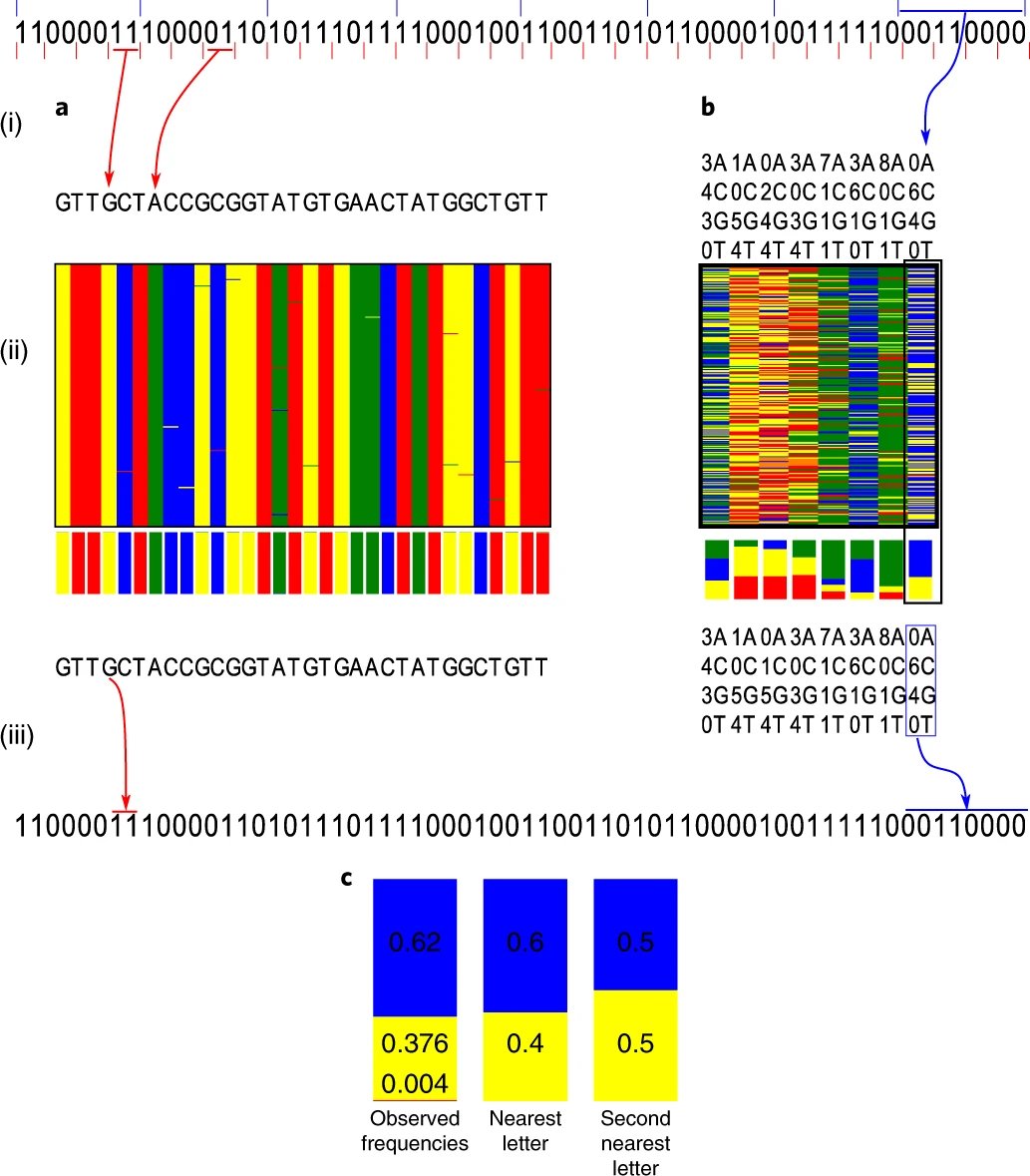
A composite letter is a mix of the original DNA letters in a position of the seq. example: 1 to 1 ratio of A and C is called M.
6/10
7/10
8/10
- Using native DNA biorxiv.org/content/10.110…
- More random access
nature.com/articles/nbt.4…
- Enzymatic synthesis
nature.com/articles/nbt.4…
nature.com/articles/s4146…
- Fast reading using nanpore
nature.com/articles/s4146…
and many other open questions
9/10
10/10


