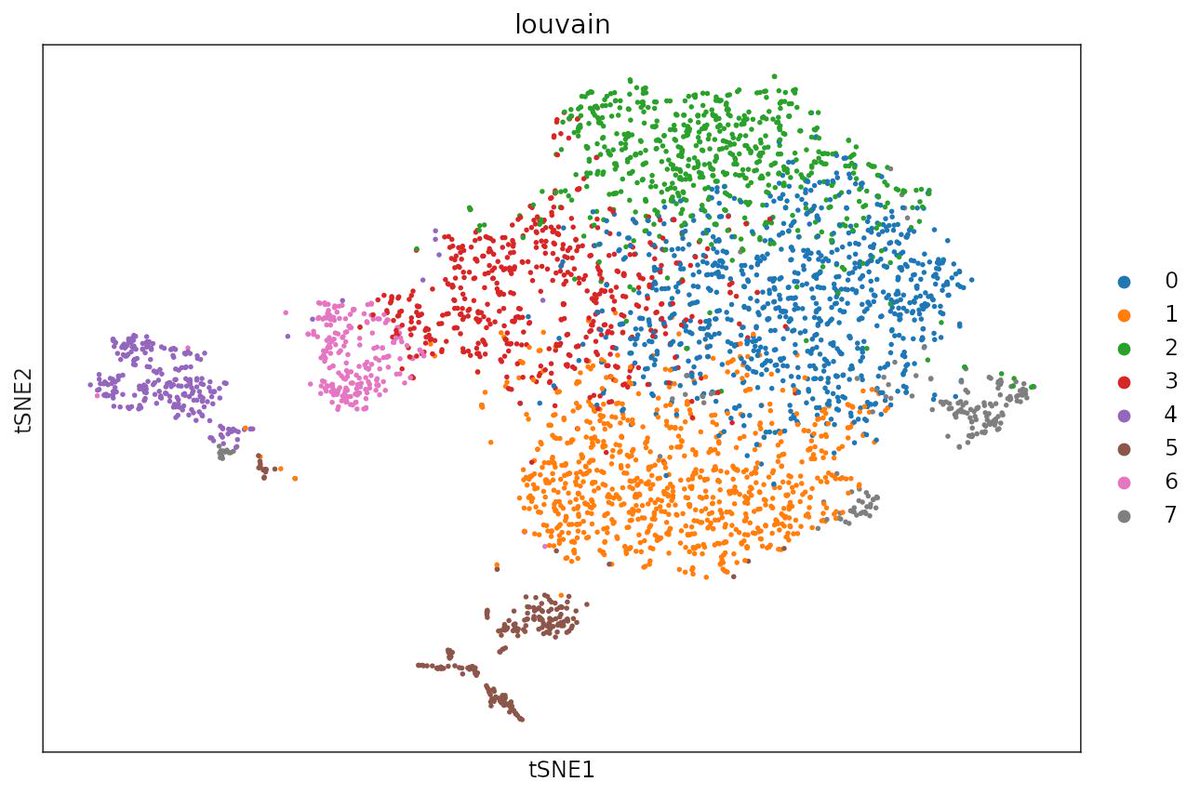
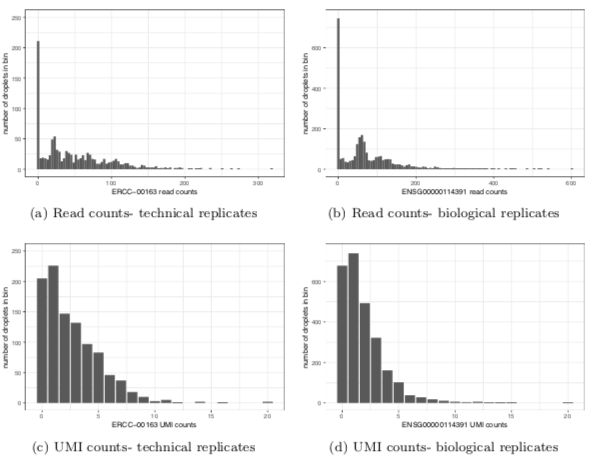
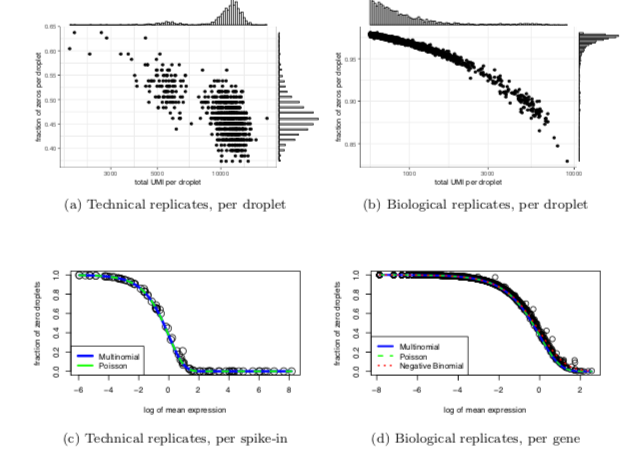
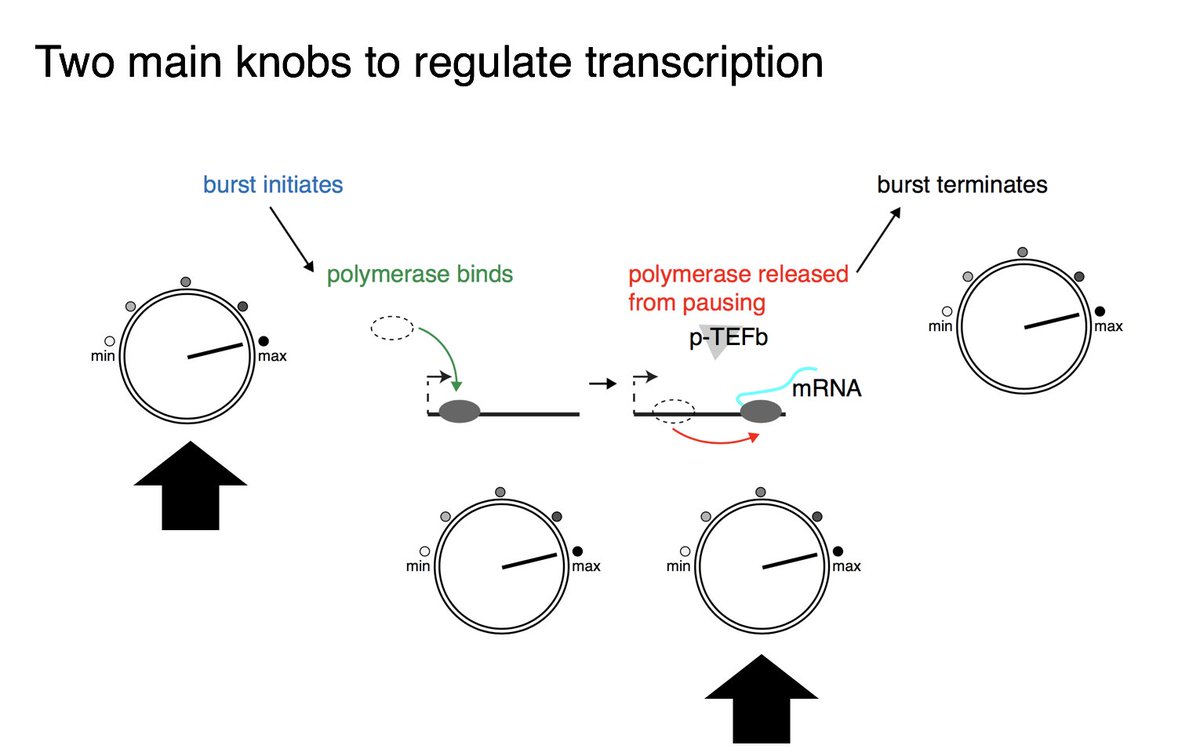
How clean is too clean?
Presenting...Molecular Cross-Validation
biorxiv.org/content/10.110…
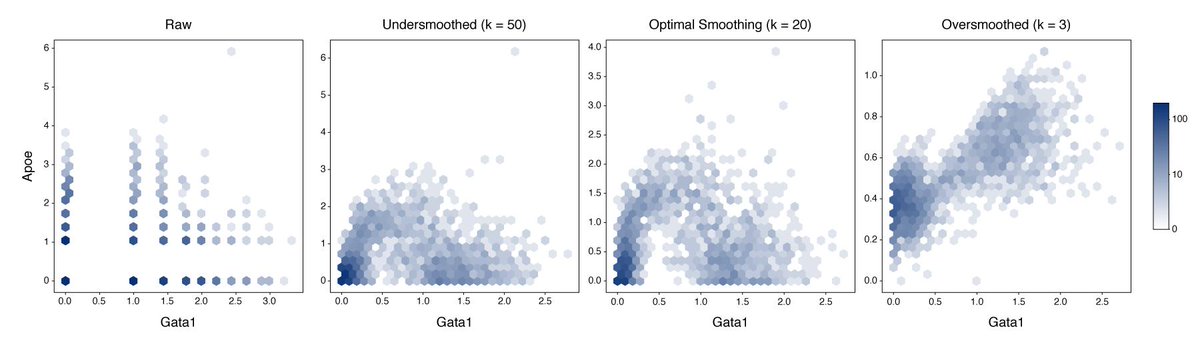
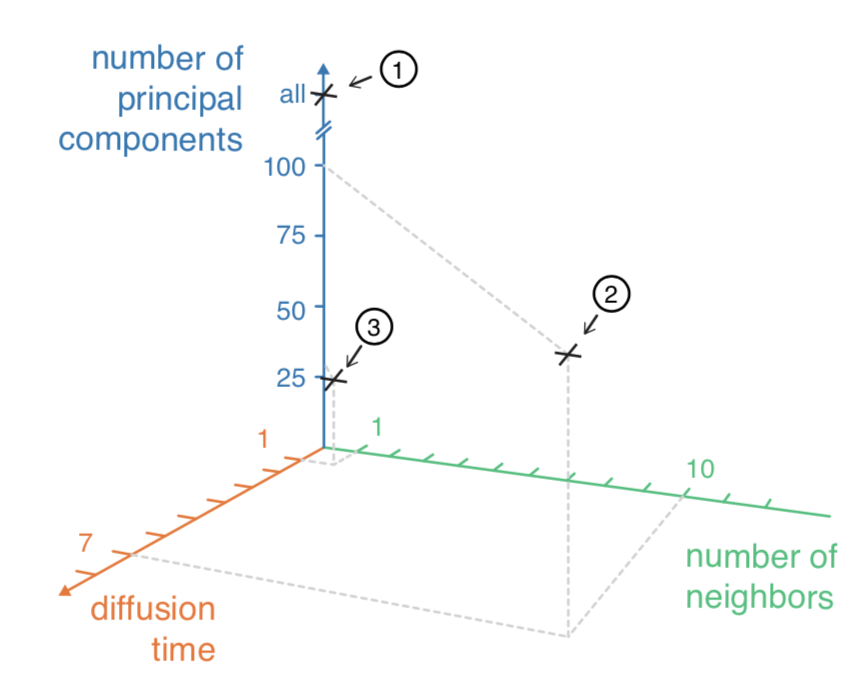
How big do you make the neighborhood? How long do you diffuse?
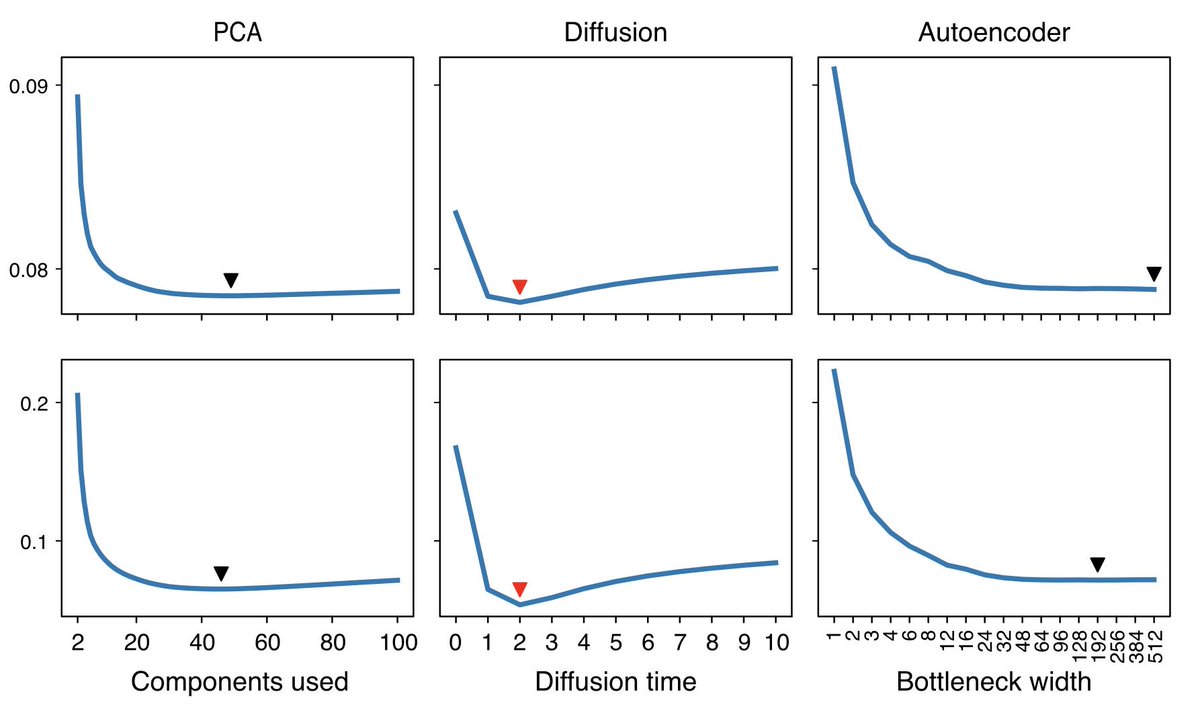
But why 20 PCs? Why not 3? Or 50?
Parameters matter here, and it’s hard to tell how tweaking things like learning rate, bottleneck width, or the random seed will affect the data.
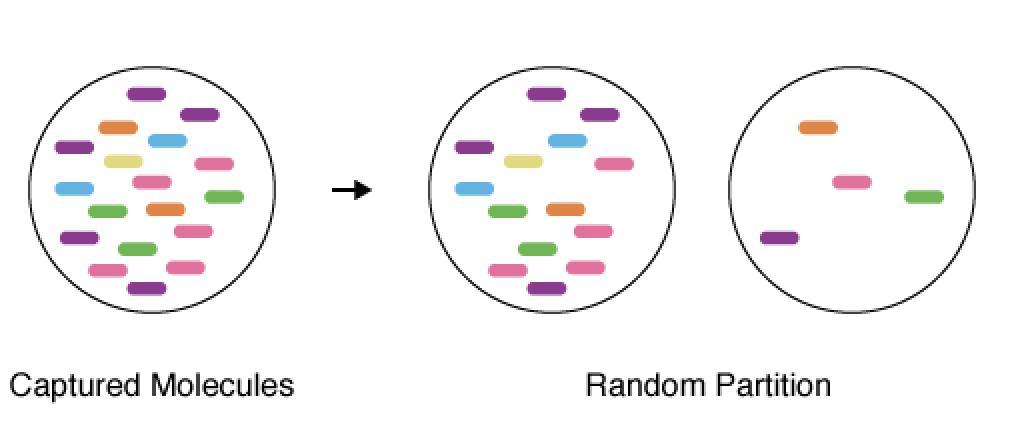
In classic cross-validation, you would split the cells into two groups, fit the model on the first group (training), and evaluate its accuracy on the second group (validation).
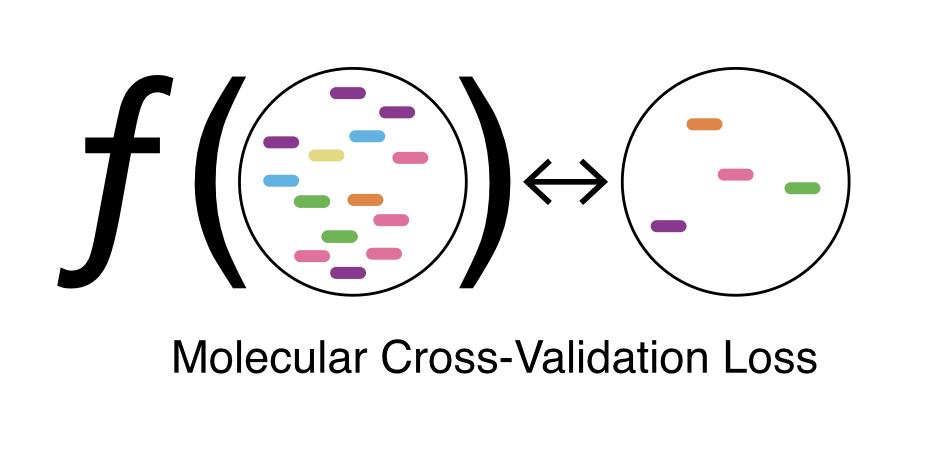
The proof is 8 lines; check out the methods section if you’re interested.

MCV lets you calibrate any model (we do PCA, diffusion, and a deep autoencoder), and pick the best one for your dataset.
If you make software which does denoising, we’d love to get MCV inside of it. Looking at you @satijalab, @theislab.
It was fun to see this story fill out and mature from the sketch in #Noise2Self, seeing how far the pixel:molecule analogy could take us.
arxiv.org/abs/1901.11365