Our nation-wide study on the #genomic #epidemiology of #SARS-CoV-2 #superspreader_events in #Austria is out in @ScienceTM !
doi.org/10.1126/scitra…
Please find the highlights in this thread. 1/8
doi.org/10.1126/scitra…
Please find the highlights in this thread. 1/8

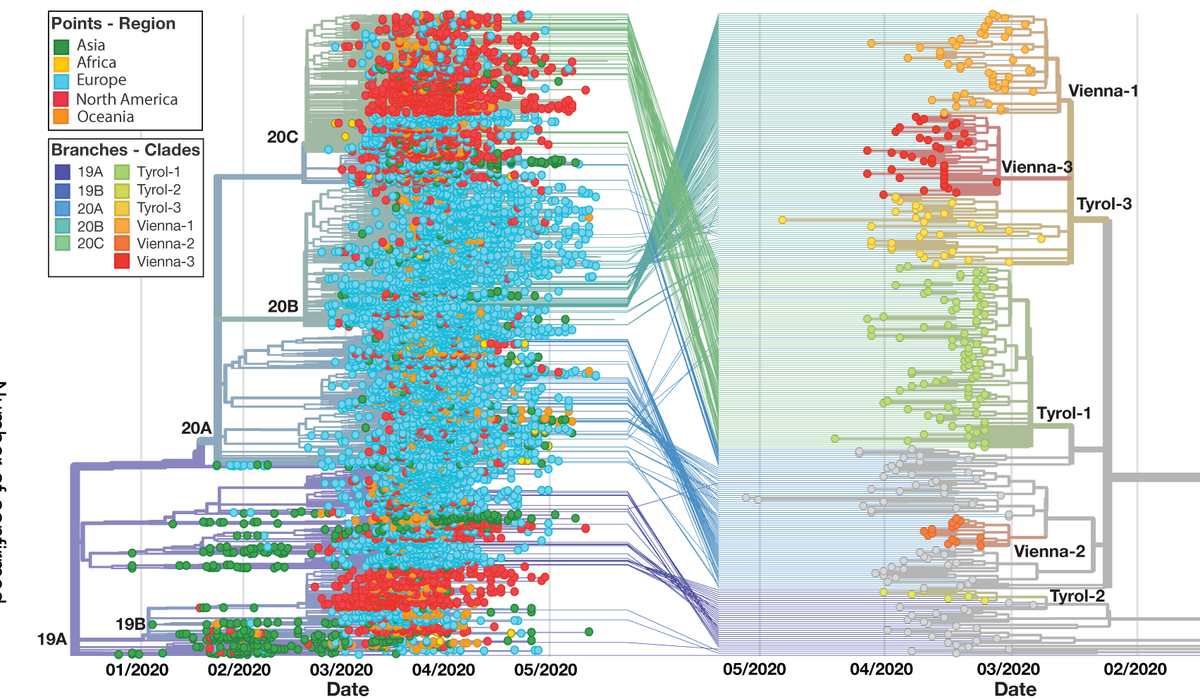
Through #Phylogenetic #SARSCoV2 analyses and data integration with @GISAID @nextstrain , we unveiled the links between #Austrian clusters to the global #pandemic. Explore our data and data-driven narratives at #NextstrainAustria sarscov2-austria.org ! 2/8 

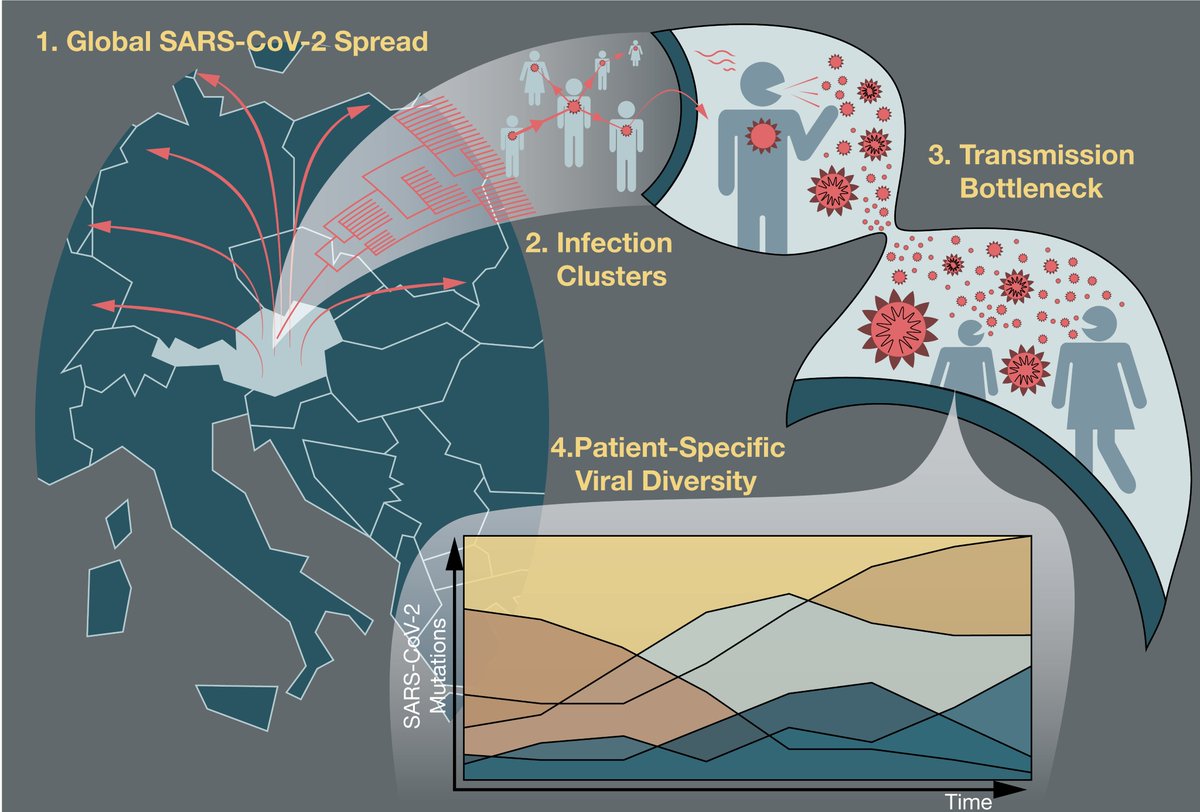
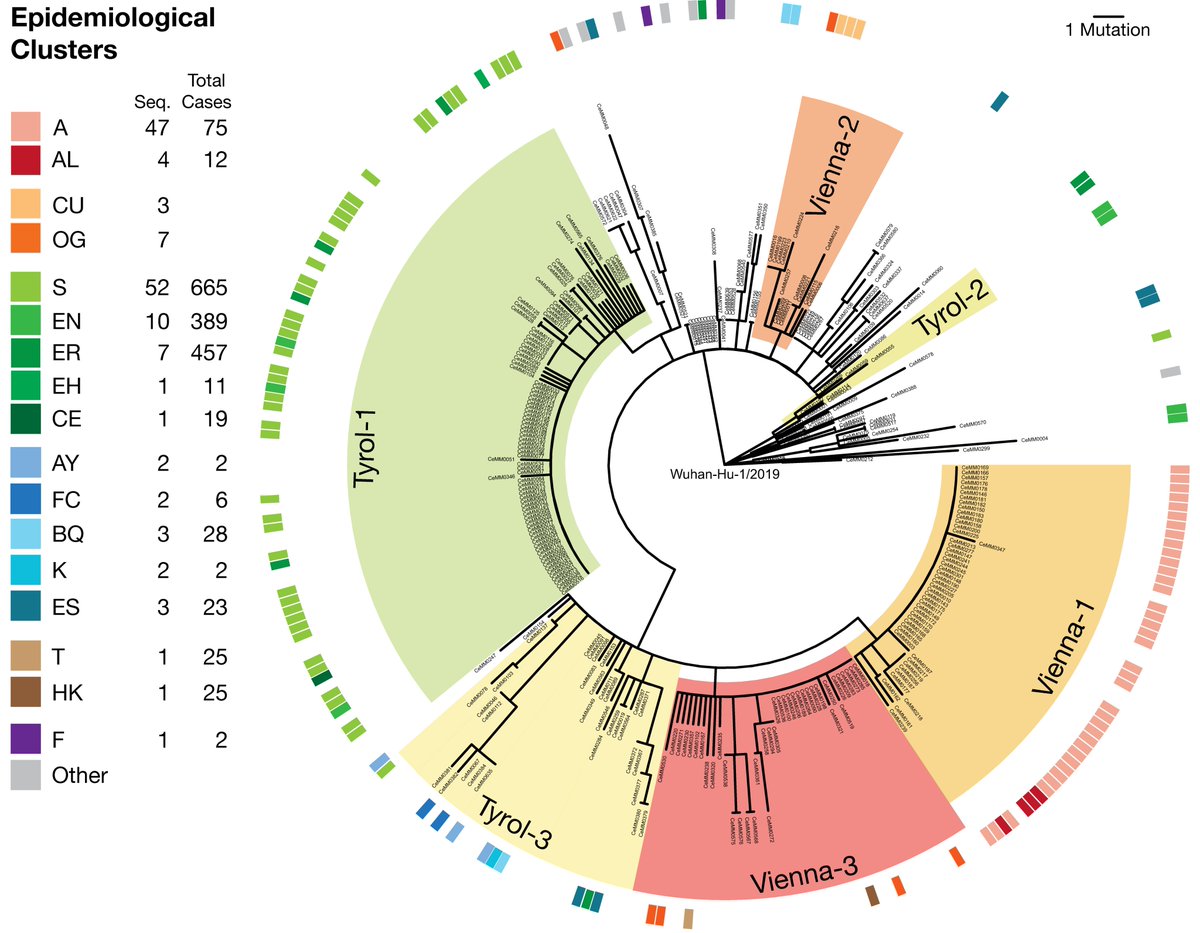
#Epidemiological and #Phylogenetic data revealed the geographic origins and distributions of SARS-CoV-2 in Austria. Different viruses circulated in the ski-resort region of #Ischgl with the most dominant #Tyrol-1 linked to @nextstrain clade 20C. 3/8 

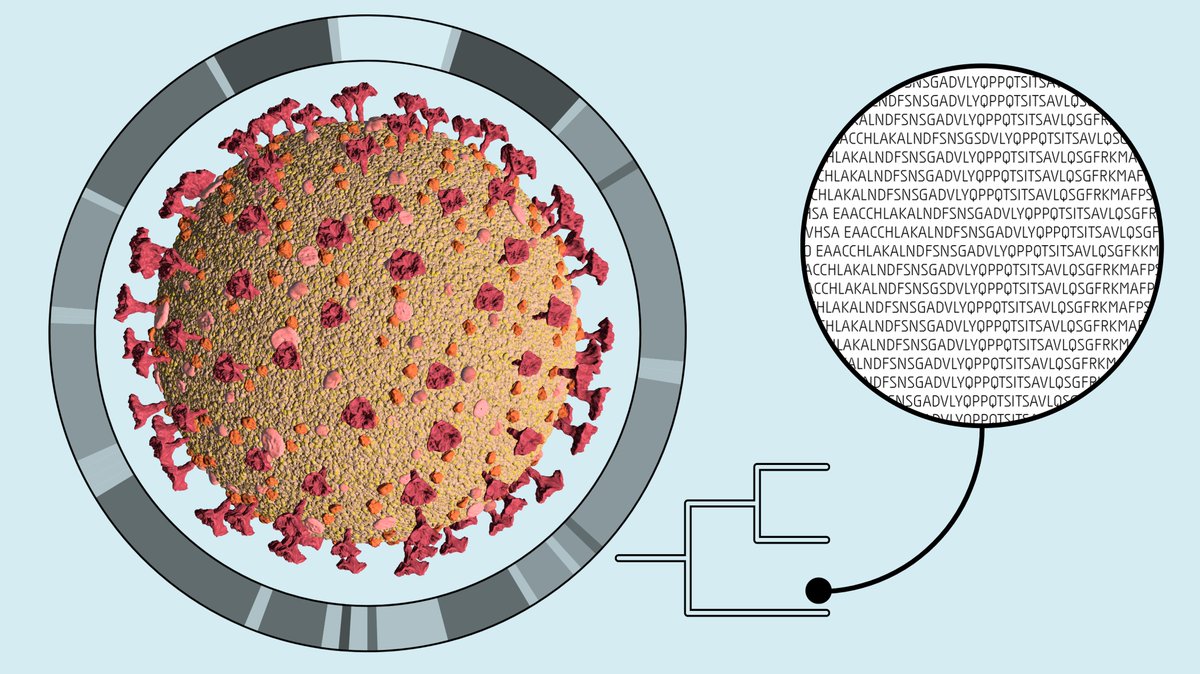
We observed the birth of a new mutation. With high-coverage #sequencing of the #SARSCoV2 genome we identified low-frequency #mutations. This enabled us to see the rise of a new #mutation and its fixation in the epidemiologically-confirmed #transmission chain of events. 4/8 

Using epidemiologically-confirmed #transmission chains and a #biomathematical approach, we determined that on average 1000 #viral particles are transmitted across infector-infectee pairs leading to a successful #infection. 5/8 

Longitudinal samples from 31 individual #COVID19 patients reveal #viral intra-host time diversity with #mutations fluctuating in frequency over time. 6/8 

Our work demonstrates the power of integrating deep viral #sequencing and #epidemiological data to better understand how #SARSCoV2 evolves and spreads through populations. This combined strategy will support future #pandemic control. Find more info: sarscov2-austria.org 7/8 
We thank our collaborators from @MedUni_Wien, @agesnews, @MedUniInnsbruck, @Harvard, @DanaFarber, @univienna, @VetmeduniVienna, @IRBBarcelona, Kaiser Franz Josef Hospital and @broadinstitute for their great efforts. Thank you to @oeaw and @WWTF for support and funding. 8/8 

• • •
Missing some Tweet in this thread? You can try to
force a refresh