New insights into SARS-CoV-2 immune evasion
#SARS-CoV-2 #mutations can lead to antibody #escape. What about T cells? In our study in @SciImmunology we report how mutated epitopes evade T cell responses.
immunology.sciencemag.org/content/6/57/e… 1/7
#SARS-CoV-2 #mutations can lead to antibody #escape. What about T cells? In our study in @SciImmunology we report how mutated epitopes evade T cell responses.
immunology.sciencemag.org/content/6/57/e… 1/7

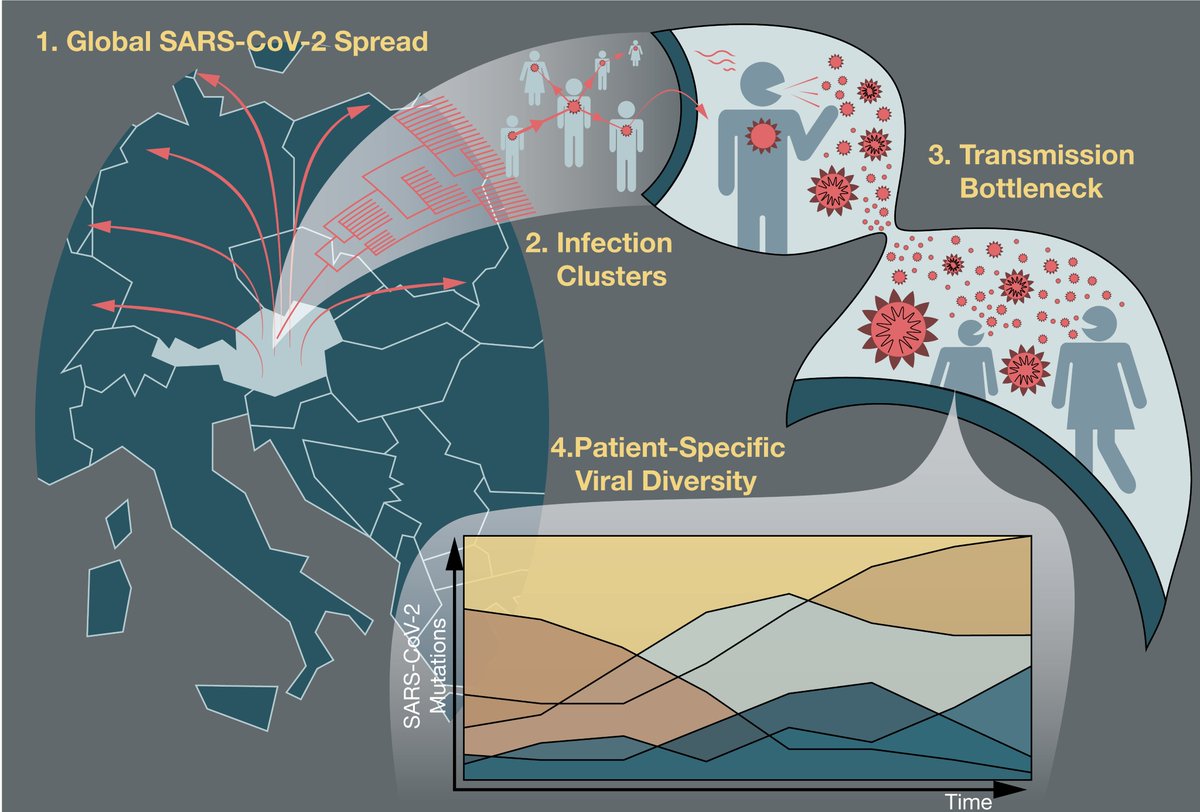
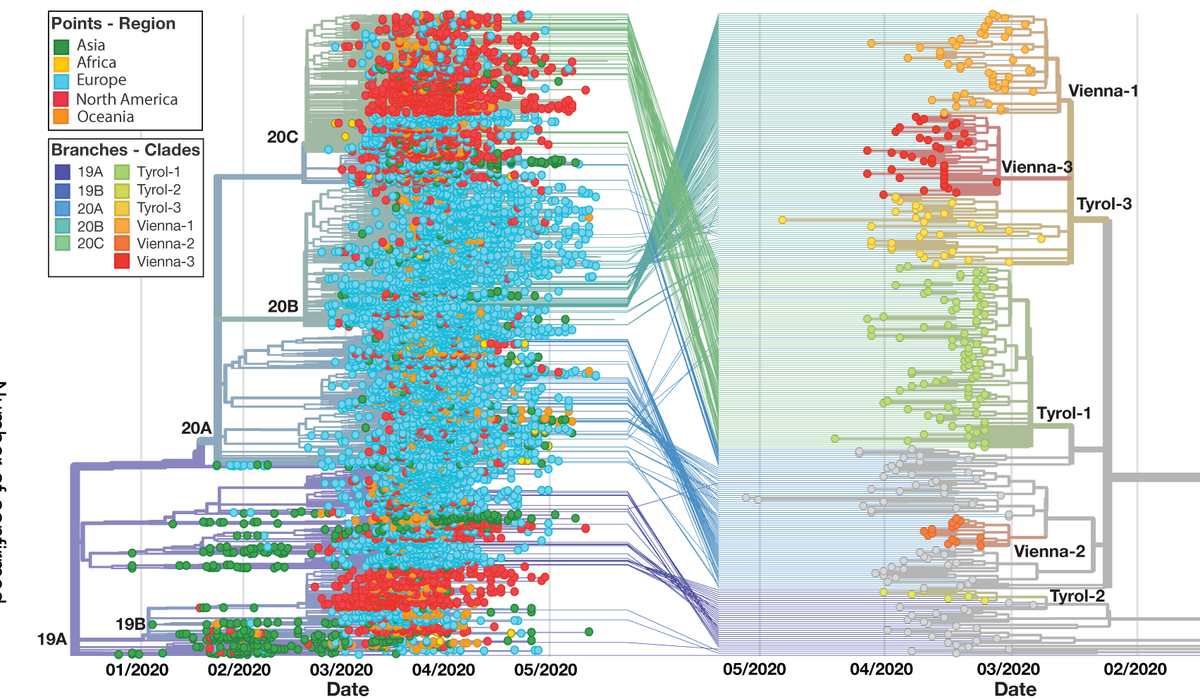
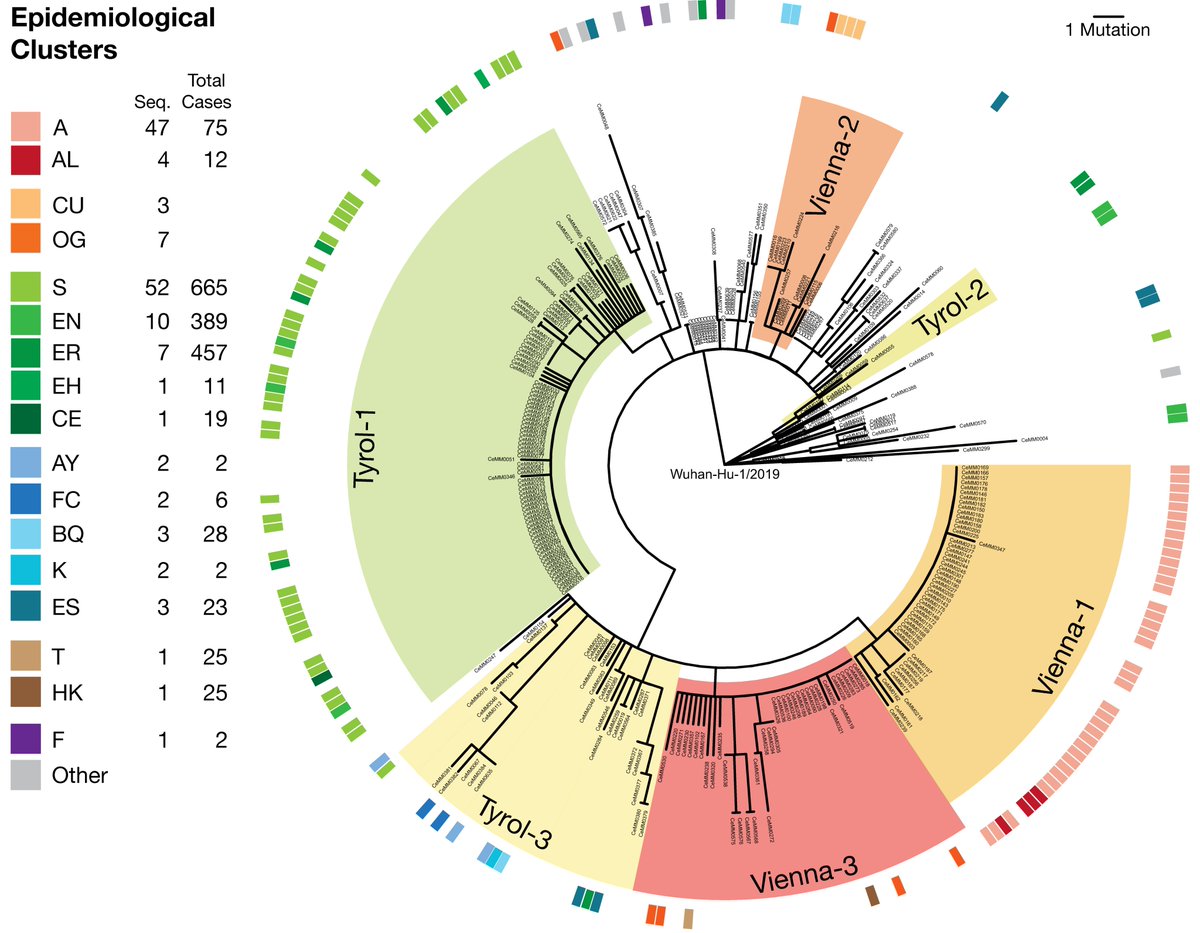
Using deep viral sequencing we identified low-frequency mutations in experimentally validated CD8 T cell epitopes of SARS-CoV-2, some of which were found as fixed mutations in global virus sequences from @GISAID. 2/7
Guided by binding predictions we selected epitopes and #mutants for further biochemical investigations by cell-free MHC-I binding assays. These analyses suggested that single amino acid changes impair binding to MHC-I molecules. 3/7 

PBMC experiments from #COVID-19 patients highlighted a strongly impaired CD8 T cell response towards mutant viral peptides. 4/7 
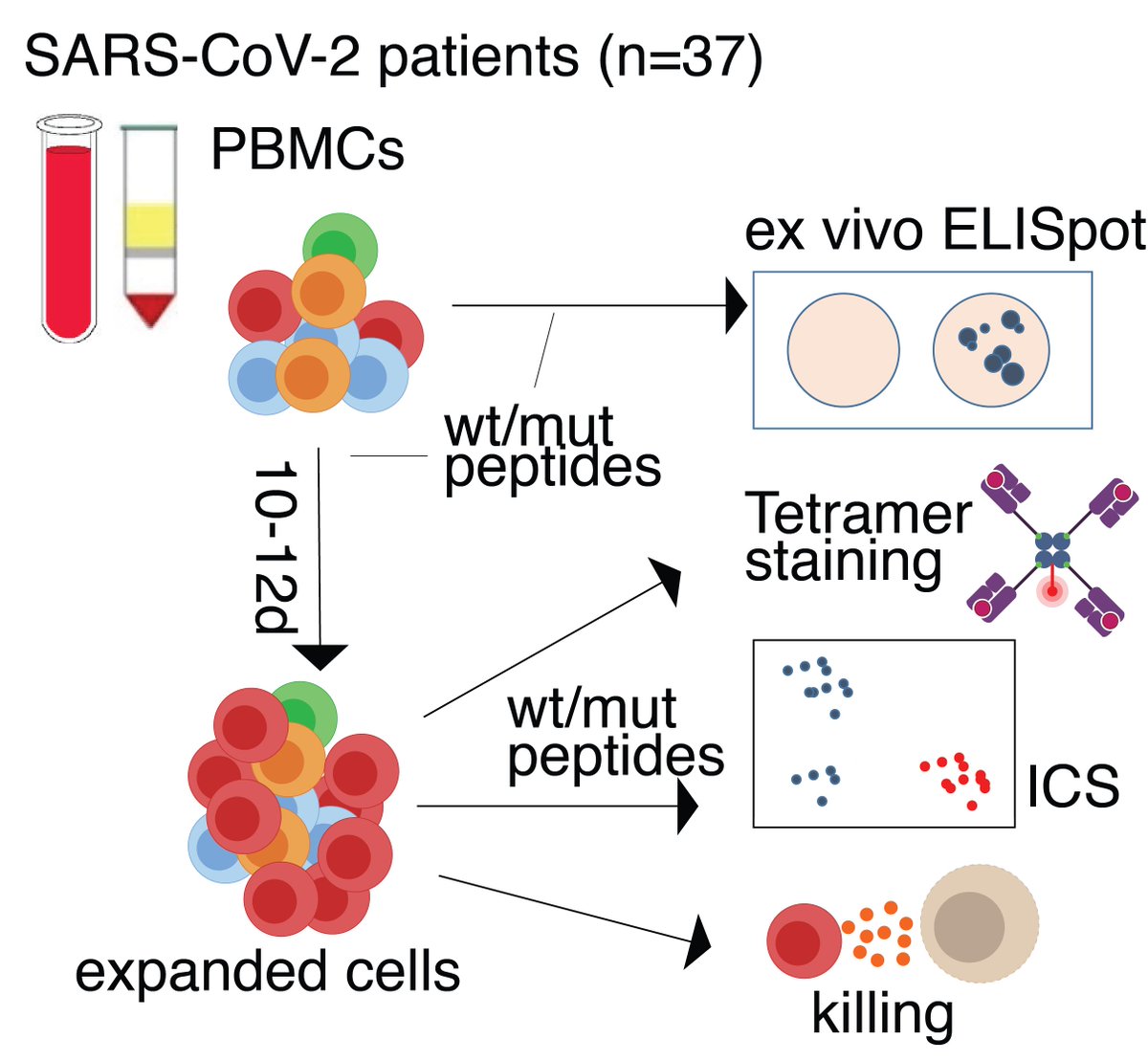
This was confirmed and extended by #scRNA-seq and TCR clonotype analysis. 5/7 
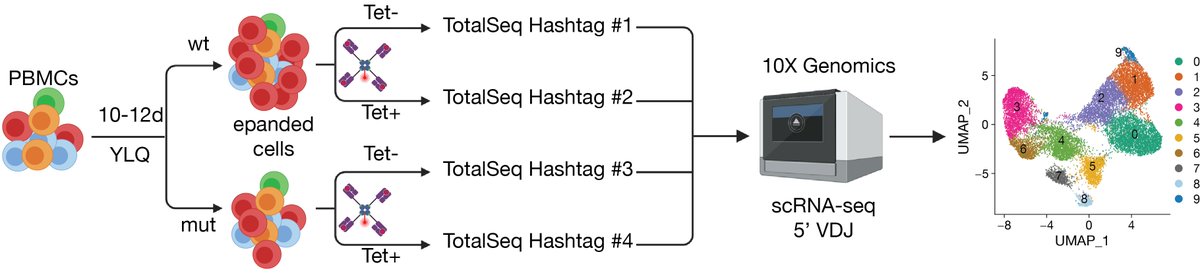
Such epitope mutations may impact T cell responses with current #vaccines that only include the viral S gene. This is yet another reason for close monitoring of emerging viral mutations and to develop vaccines inducing broad immune responses. 6/7 

We thank all #collaborators from @CeMM_News @oeaw, @MedUni_wien, @StAnna_CCRI, @Wiengesundheit, @MedUniInnsbruck, @unibern and @agesnews for their contributions. Thank you to @oeaw, @wwtf, @fwf_at and @ERC_Research for support and funding. 7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh