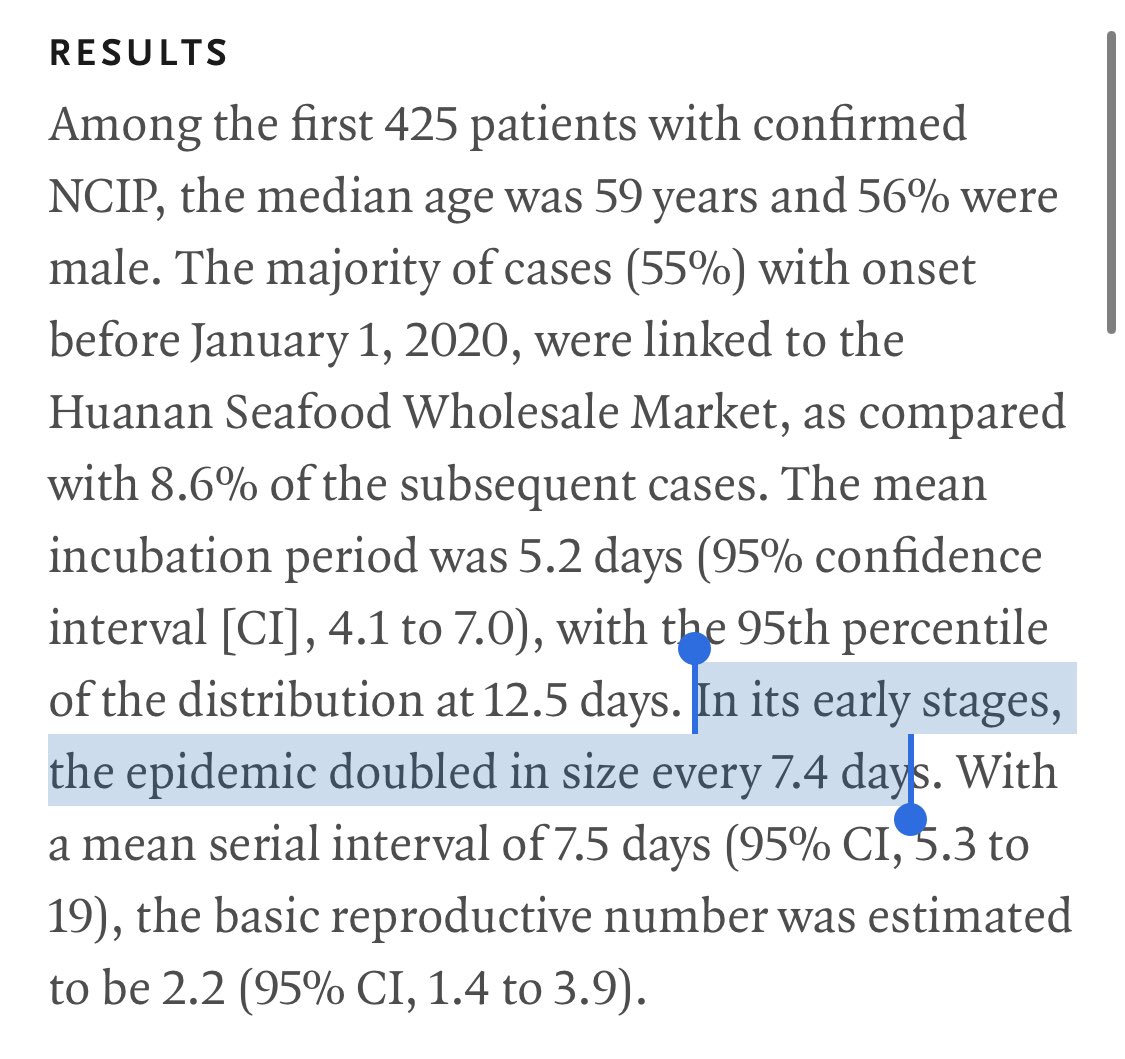
But if it did work, you might be able to let the subgroup with the lowest predicted risk out of quarantine after genotyping them.
This can unblock the production of vaccines, antivirals.
Knowing who's at higher risk may help a lot here.
There's a literature on genetic determinants of infectious disease susceptibility. Some cites:
1) Malaria: ncbi.nlm.nih.gov/pubmed/2289518……
2) Theory:
ncbi.nlm.nih.gov/pubmed/2372490……
3) 20+ diseases: ncbi.nlm.nih.gov/pubmed/2892844……
4) 10+ diseases:
ncbi.nlm.nih.gov/pubmed/30053915


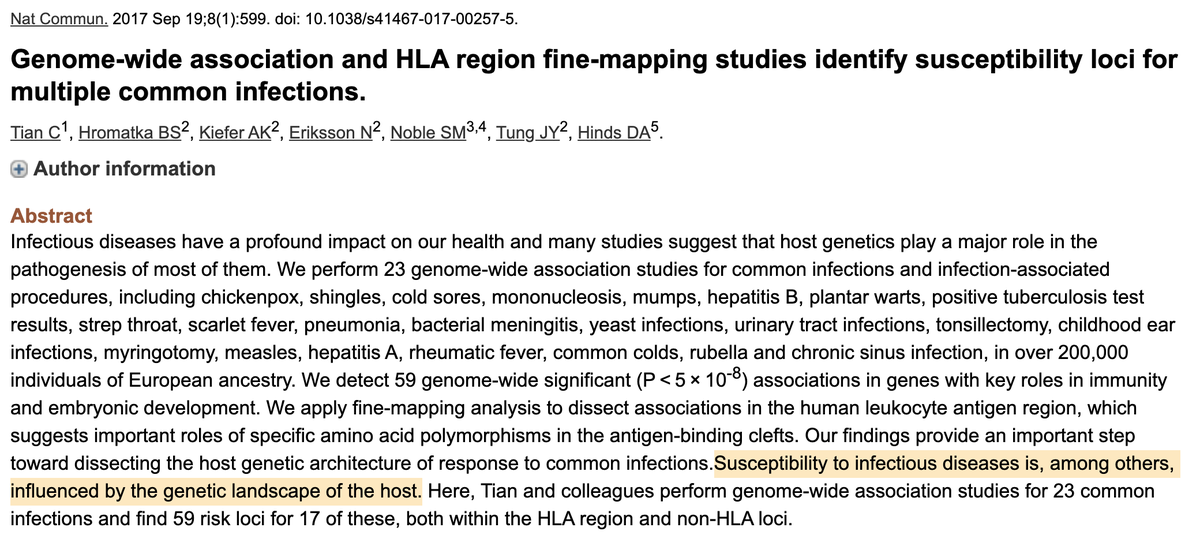

Sometimes you get great signal from just a single variant, or a small number (eg OCA2/eyes). Other times you need many (eg PGS).
So this would have to give strong signal. But the ROI is high if it works.
2) Specific inputs are flexible. As noted in OP, can be phenotype + genetics.
3) Age/gender: cheap to ascertain, high value. Assess other predictors via ascertainment cost vs predictive benefit.