nature.com/articles/s4158…
Several points worth emphasizing from this paper.
Twitter explainer below 👇, also great N&V piece by Wray & Gratten here:
nature.com/articles/s4158…
1/15
Particular congrats to @DonnaWerling, Harrison, @joonomics, @m_sto, and Lingxue, who spearheaded the analyses from start to finish!
Now, on to the science...
2/15
As such, WGS association studies must be careful about multiple testing burden and adjust p-values accordingly.
3/15
4/15
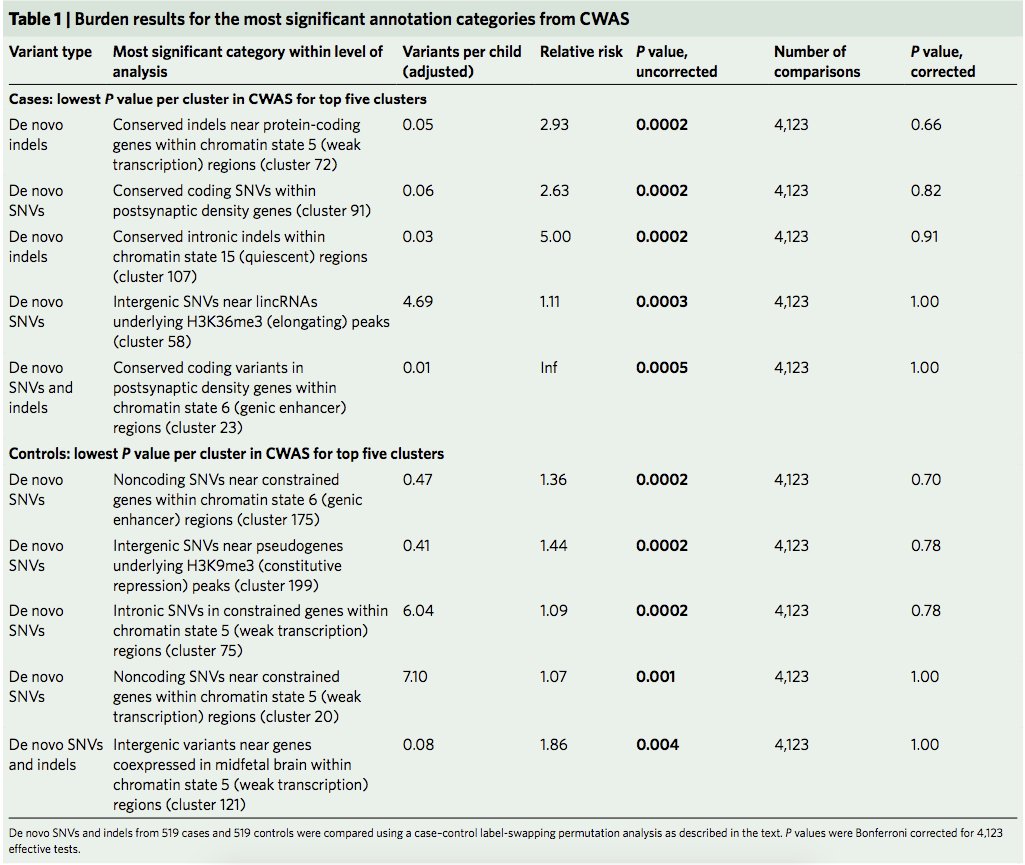
Most signif. in cases: DNVs in conserved bp within postsynaptic density genes
Most signif. in controls: noncoding DNVs in genic enhancers near constrained genes
5/15
6/15
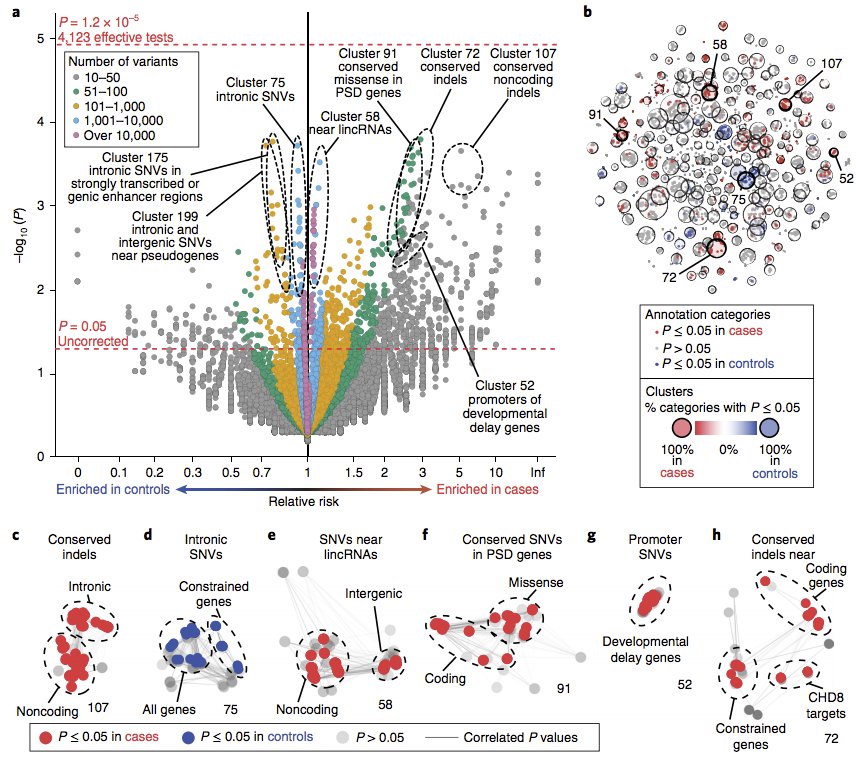
Meet the category-wide association study (CWAS) -- great work from Devlin & Roeder labs
7/15
8/15
9/15
10/15
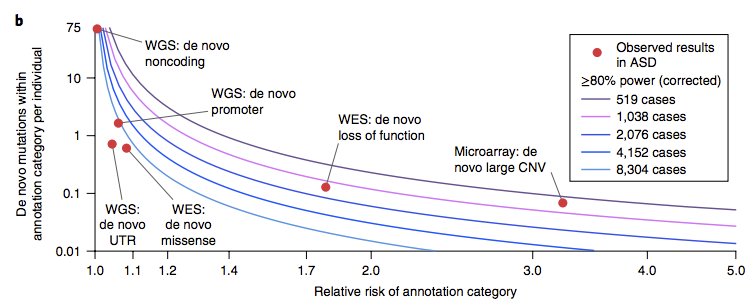
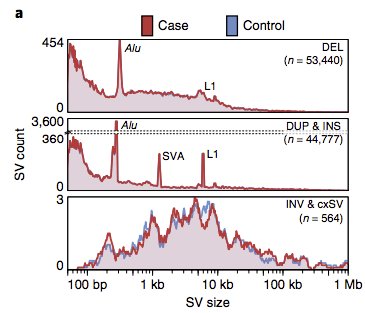
We developed an SV pipeline that yields close to 6k SV (>50bp) per genome at FDR < 1%.
>5k high-quality SV can be routinely captured from garden-variety 30-40X Illumina WGS.
11/15
12/15
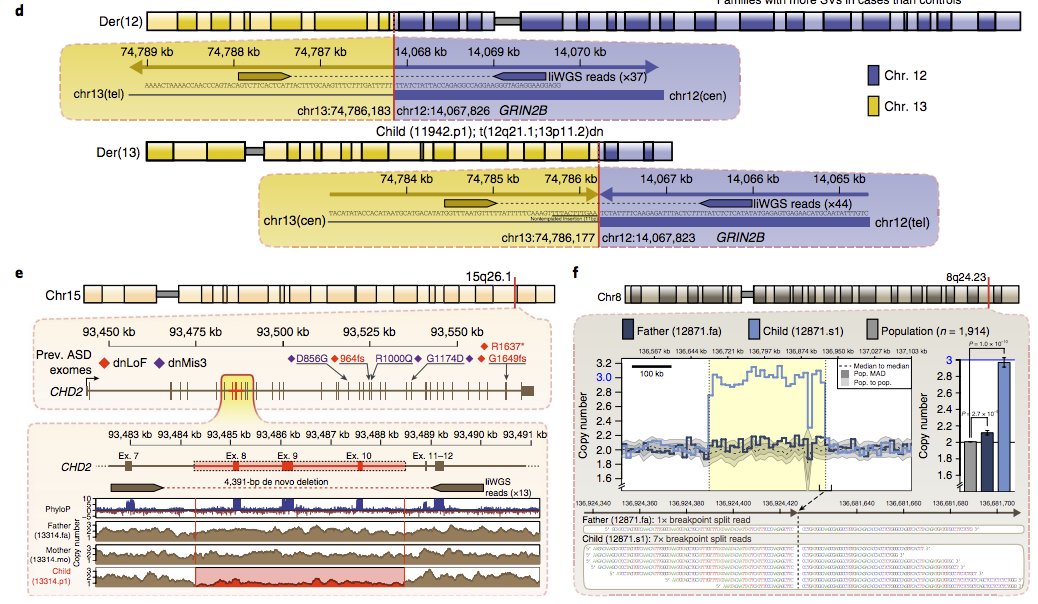
Some are even likely relevant to autism, like a de novo balanced translocation of GRIN2B or a de novo 4-exon CHD2 deletion.
13/15
That said... please, please treat significant noncoding associations from small studies with skepticism until multiply replicated in larger cohorts.
14/15
Was awesome to work on this over last ~3 years. Great group of collaborators! Looking forward to much more WGS soon.
~Fin~
15/15