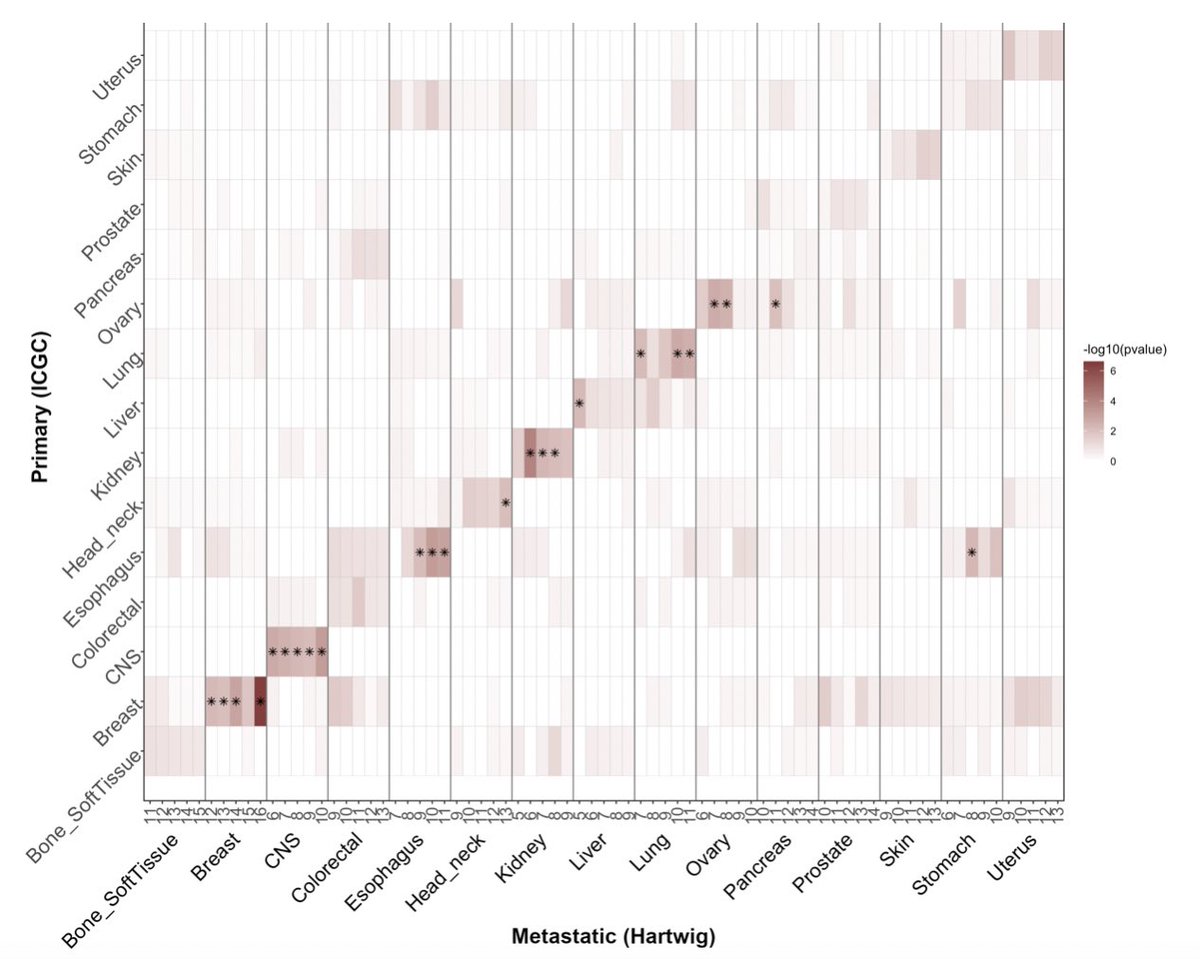
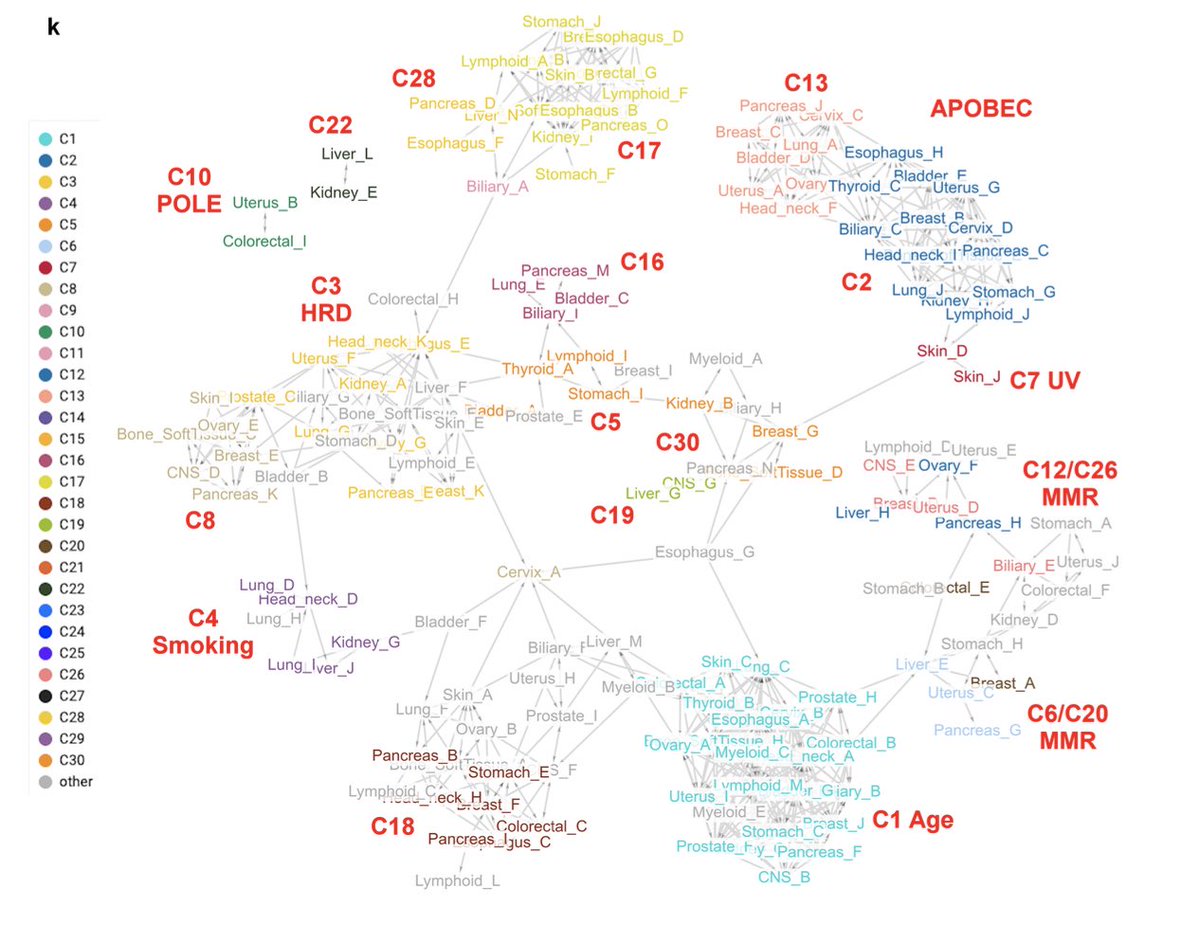
Showcasing the complexity of #mutationalsignatures
A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies @NatureCancer
👇
1/ nature.com/articles/s4301…
Try the browsable map here:
signal.mutationalsignatures.com/explore/cancer…
5/
And perform signature assignments on your samples!
@SignalCambridge
signal.mutationalsignatures.com
14/
Code: github.com/Nik-Zainal-Gro…
Special Kudos 👍💪 to #AndreaDegasperi #ScottShooter #JanCzarnecki for driving this project.
Though the whole team👇
were involved really!
/END