cis-eQTLs and splicing-QTLs reveal mechanisms by which a DNA variant can impact mRNA abundance. It's good to model and predict these.
At a particular locus these may or may not translate into elucidation of the causal transcript for the disease phenotype.
One example is the glucokinase gene (GCK) which sits near a diabetes SNP and is also a MODY gene
type2diabetesgenetics.org/gene/geneInfo/…
omim.org/entry/138079
nature.com/articles/nrg.2…
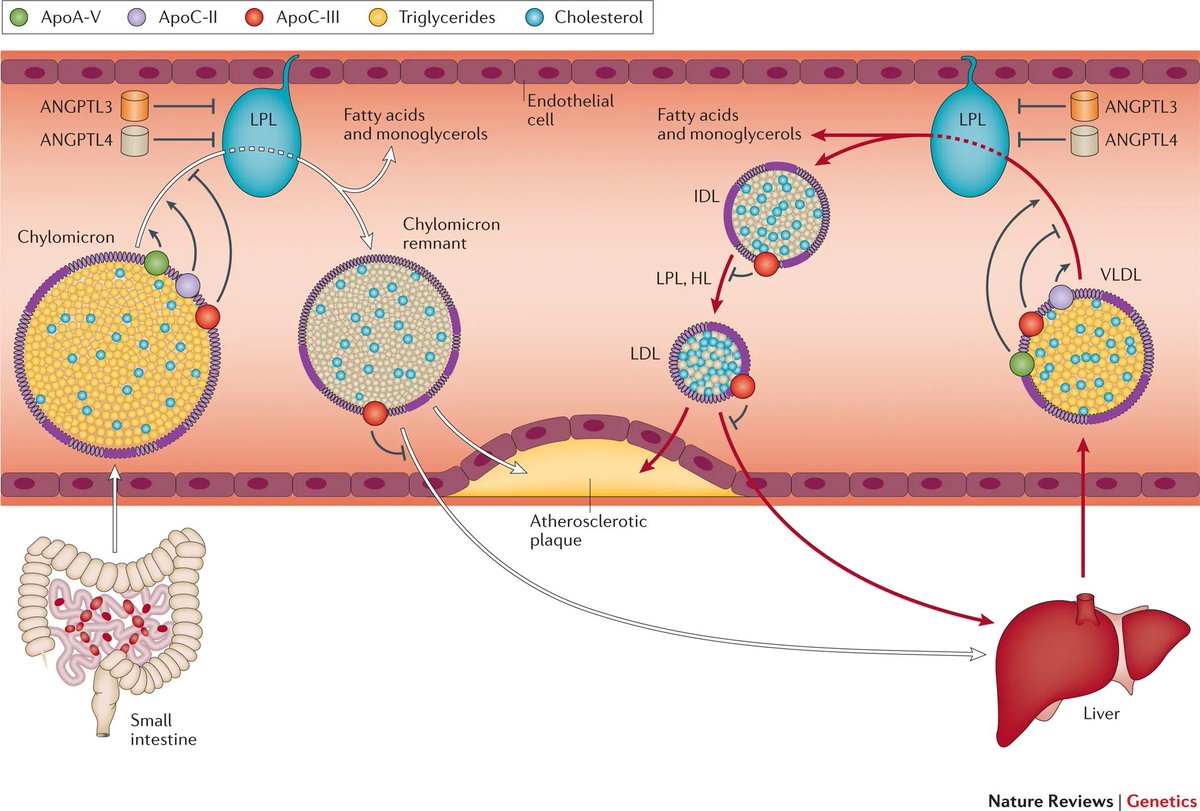
genetics.opentargets.org/variant/5_7536…
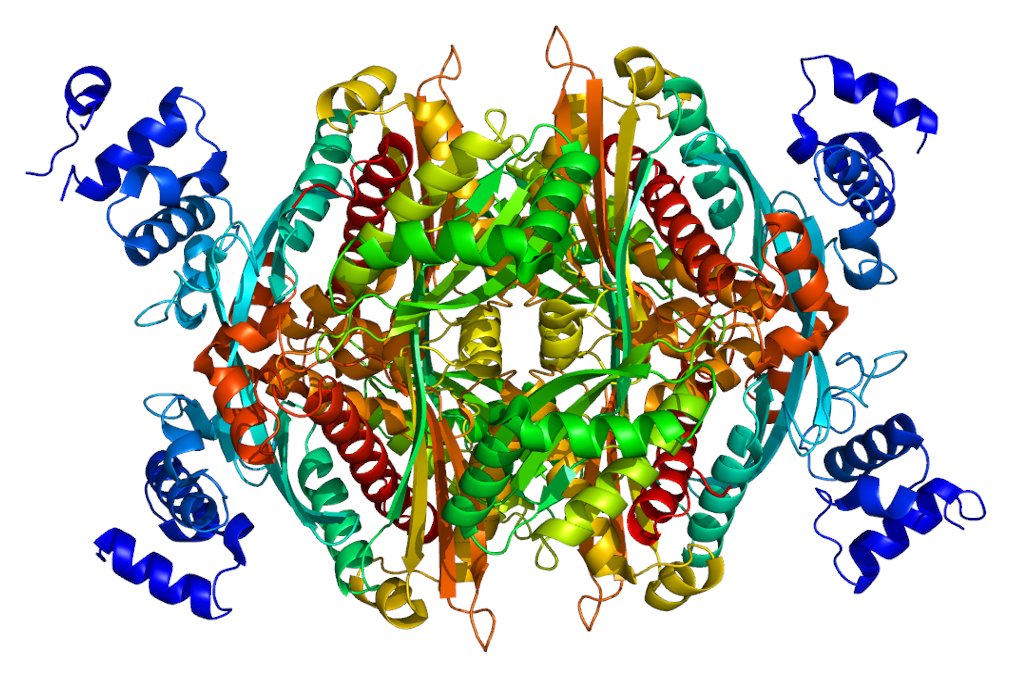
Eg: rs1558902 for BMI acting through IRX3/IRX5, Claussnitzer, 2015
ii) rs7528419 for LDL-C acting through SORT1, Musunuri, 2011
Are there categories of "gold standard SNP->gene" examples I've missed?
Do you disagree with any of the buckets I've presented here?










