But first an intro to what open science is...
@daniellecrobins and @rchampieux have this great umbrella infographic!
[Just need to video call a student to explain some cog modelling I did for their exp! BRB!]
Time to get back to this thread and link it up to what I wanted to get to — before I have to run off again for a talk! 😂
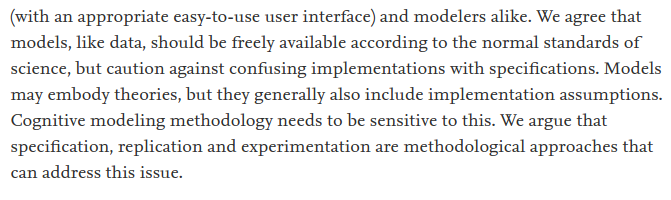
Now all the intro stuff is out of the way... #openscience in part aims to help scientists make their data and code open.
1) coding up the experiment itself, the data collection is done using some kind of program, e.g., check out @psychopy;
This last bit of coding is the kind of work I mostly do, which has a large number of people often replicating other people's work as part of the learning process.
Well...
But how? 😕
Please feel free to ask any questions!
doi.org/10.1016/j.cogs…
doi.org/10.7717/peerj-…

More here: oliviaguest.com/doc/guest_roug…




