illumina.com/systems/sequen…
Below is a general overview of a whole genome sequencing workflow (sample in, data out).
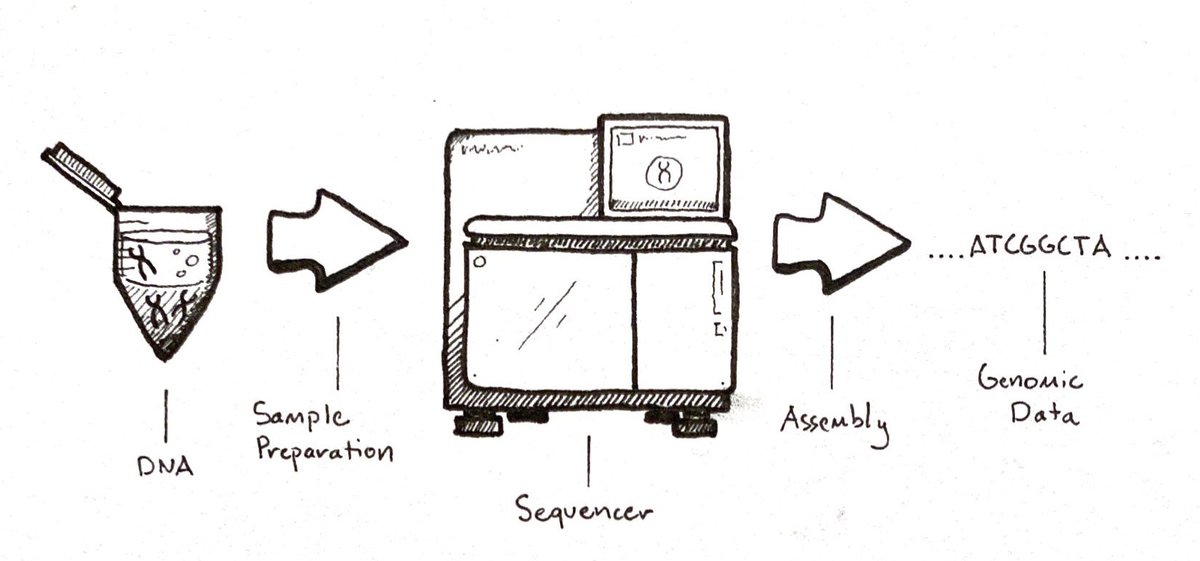
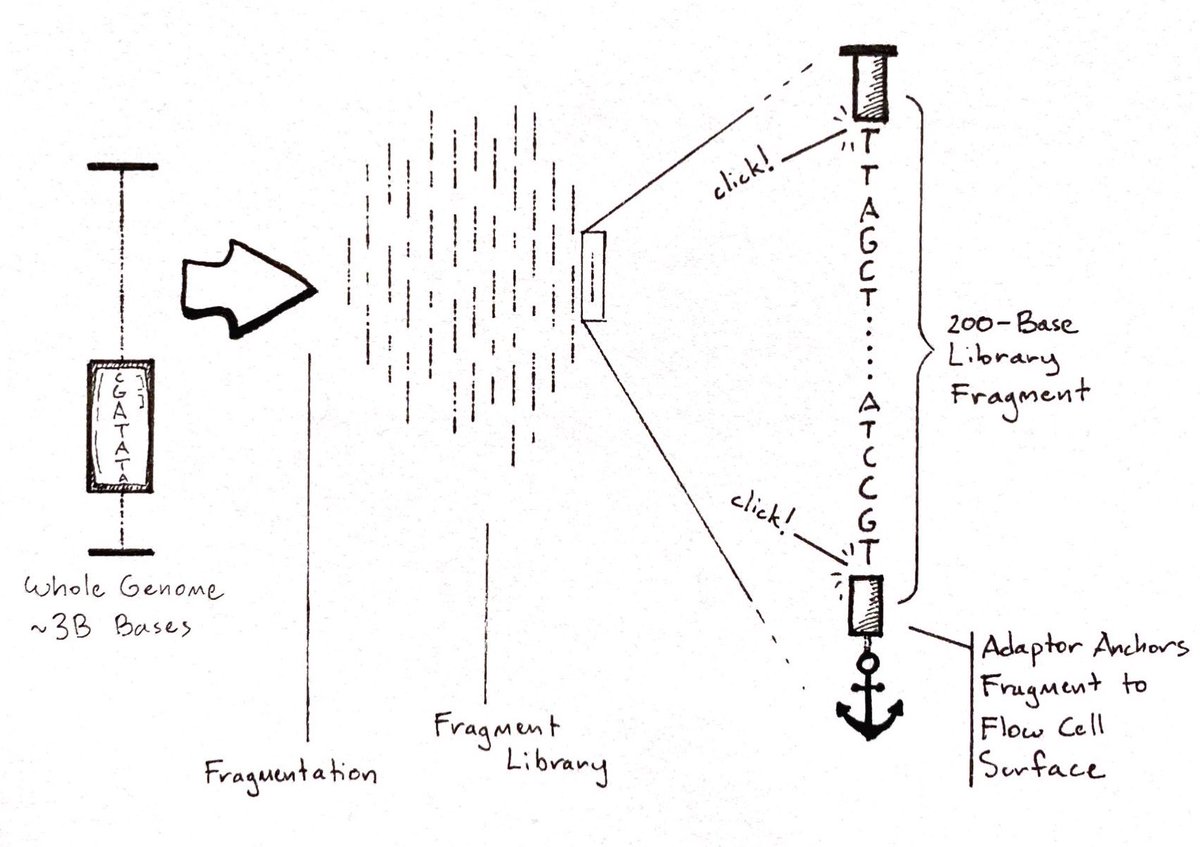
Later, algorithms reassemble the pieces like a massive jigsaw puzzle.
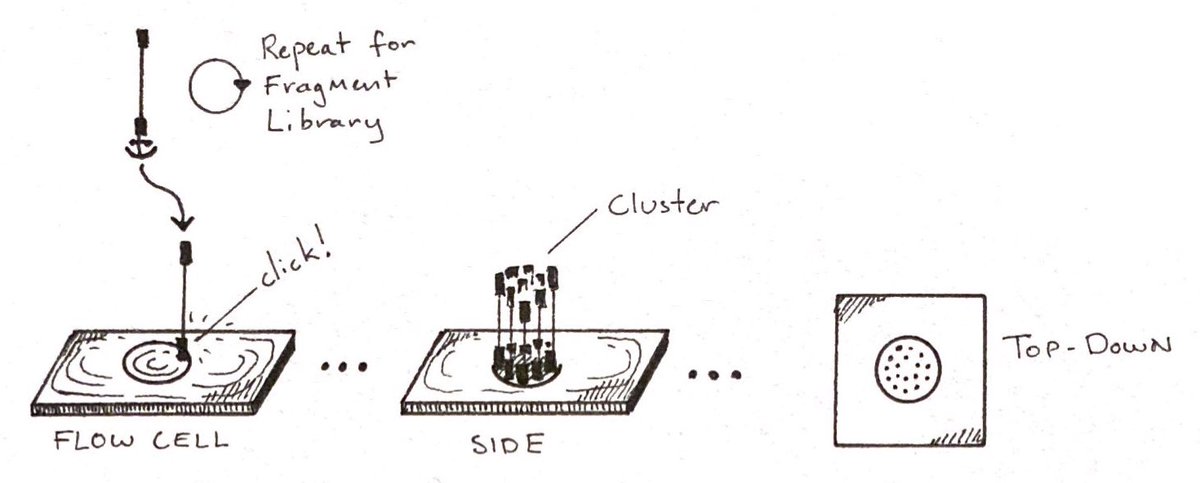
Fragments organize themselves together in uniform clusters, which makes reading easier.
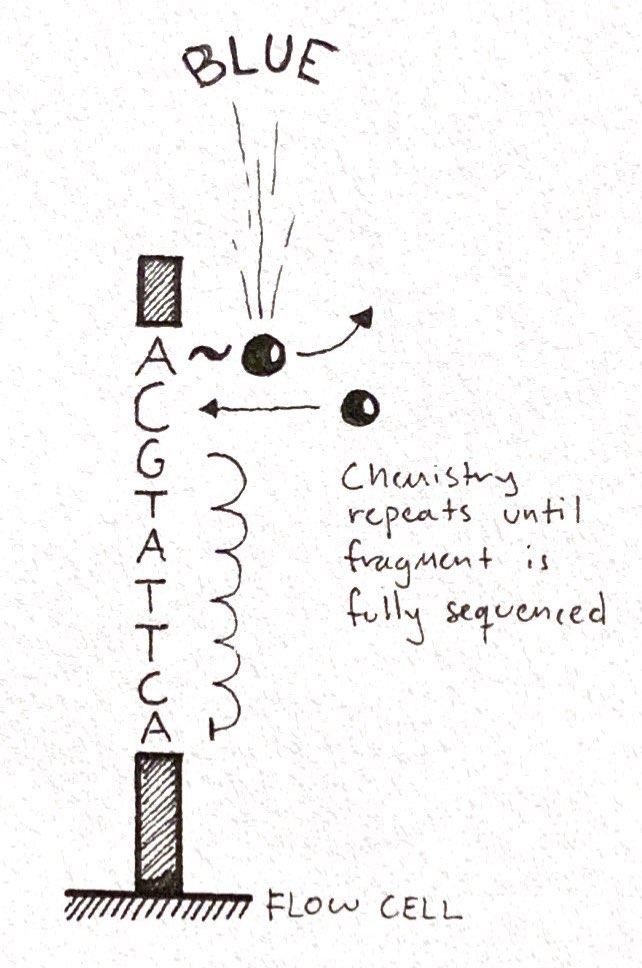
Now, let's get into the specific of the NextSeq 2000...
Tighter clustering of molecules on a flow cell = more base reads in an image/run = amount of #reagents (chemicals) required is smaller = Cheaper!
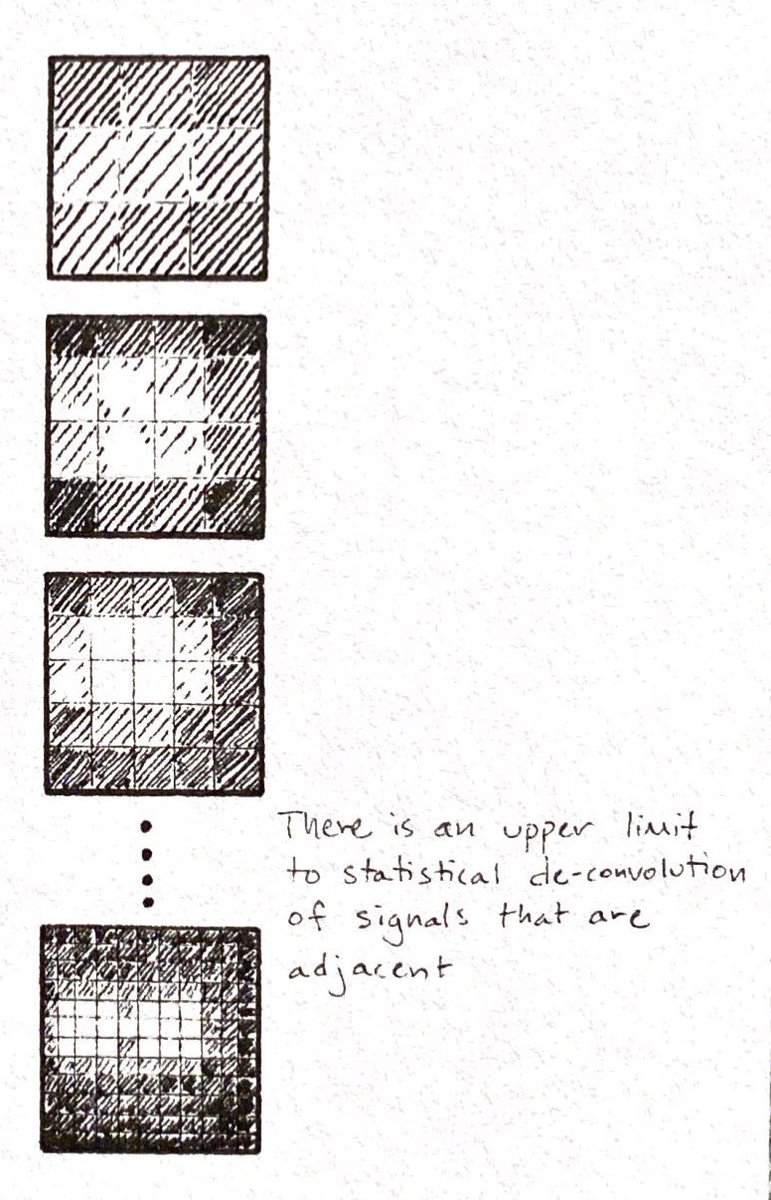
There's a limit to how tightly you can pack fragments in a cluster.
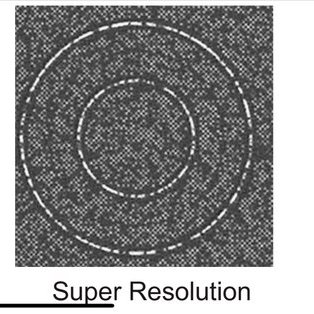
This is what super-res/new chemistry overcame...
The #NextSeq 2000 is capable of reading bases that are spaced 20nm apart, yielding super-resolution (breaking Abbe's limit). This doesn't mean a physical law was violated.
It means chemical/optical cleverness.
Probability ~~ function(x-position, y-position, time).
The blue-green/random-flash hack is a clever circumvention of a physical law imposed by the wave nature of light.
It enables 30% closer clustering and a significant reduction in reagents.
A clever, randomized blue-green chemistry and a high-resolution optical array enable DNA bases (that are closer together than the diffraction limit of light) to be spatially resolved, resulting in fewer necessary reagents in sample prep and a 2X reduction in OpEx.
This is not a recommendation to buy, sell, or hold any security. The intent is for educational purposes only and is based solely on publicly available information sourced below.
ILMN Patent:
patentimages.storage.googleapis.com/38/1d/51/9ba7d…
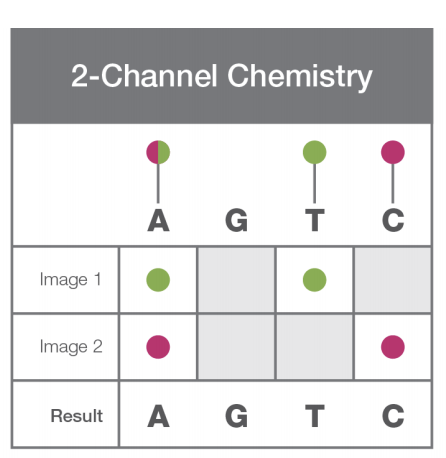
Channel Chemistry:
illumina.com/science/techno…
ILMN Slide Deck:
seekingalpha.com/article/431673…
Diffraction Limit:
microscopyu.com/techniques/sup…
Resolution Interference:
microscopyu.com/microscopy-bas…
Stochastic Photo-Switching:
microscopyu.com/techniques/sup…
Statistical Deconvolution of Images:
ncbi.nlm.nih.gov/pubmed/22677393
Excitation and Emission Spectra:
micro.magnet.fsu.edu/primer/lightan…
All Drawing My Own!