threadreaderapp.com/thread/1093179…
doi.org/10.1021/nl0500…
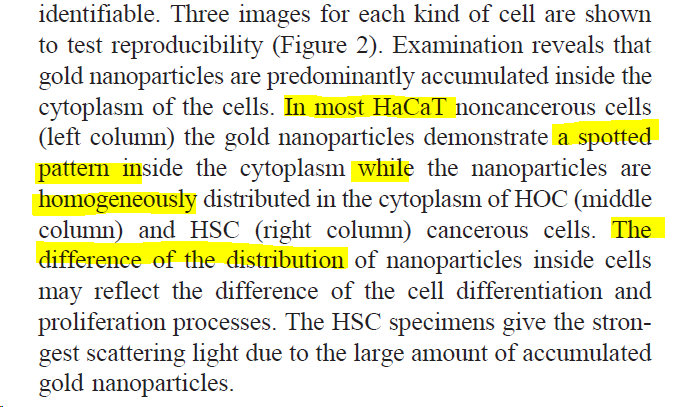
And three epithelial cell lines : one non-cancerous (HaCat) and two cancerous (HOC and HSC).
ncbi.nlm.nih.gov/pubmed/10527633
biodiversitylibrary.org/page/27095464#…


More seriously, what strikes me in this short description is how vague and unscientific the language is. It also suggests that the authors are not very familiar with looking at cells in a microscope.
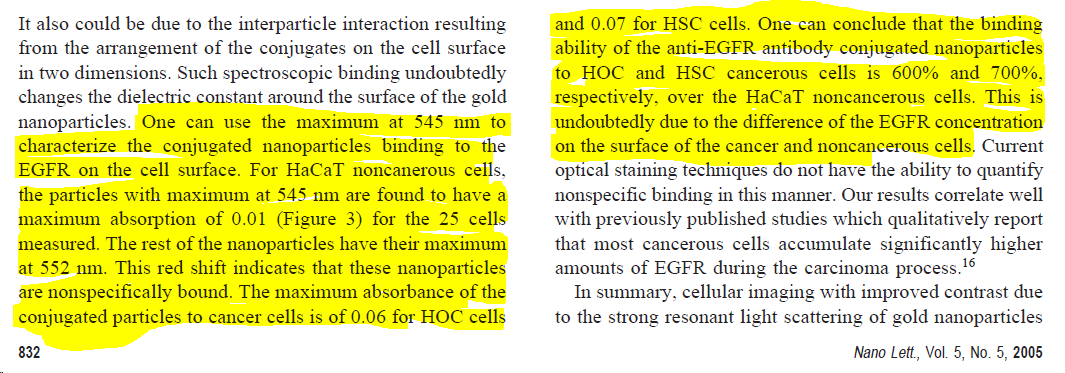
The press release, and the Georgia tech abstract, say that ".. researchers found that the gold nanoparticles have 600 percent greater affinity for cancer cells than for noncancerous cells."
Let's ignore that I can't actually see cells in Fig 3. (see 👆).
Let's also ignore that there is no affinity measurement in this paper (that would require a titration).
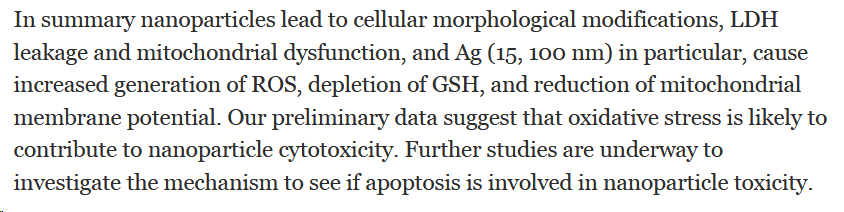
Hussain et al, 2005, In vitro toxicity of nanoparticles in BRL 3A rat liver cells.
"This study was undertaken to address the current deficient knowledge of cellular response to nanosized particle exposure."
≠ NP sizes
≠ materials silver (Ag; 15, 100 nm)
molybdenum (MoO3; 30, 150 nm)
aluminum (Al; 30, 103 nm)
iron oxide (Fe3O4; 30, 47 nm)
titanium dioxide (TiO2; 40 nm)
cadmium oxide (CdO; 1 μm)
manganese oxide (MnO2; 1–2 μm)
tungsten (W; 27 μm)
Foley et al is a single figure paper (about C60 which for a time was supposed to revolutionise drug delivery). Convinced?

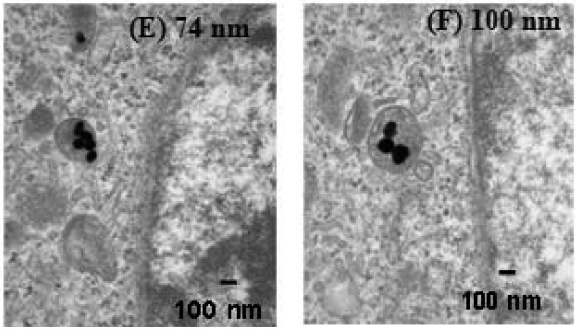
What? Where did #12 show this? Only evidence on morphological modif is for Ag NPs (fig 3) & is v poor. In any way, Ag NPs are toxic; when cell dies they do change morphology. That's due to cells dying.
"Cytotox of CdSe & CdSe/ZnS nanoparticles has been investigated 4 ≠ surface modifs such as coating w/ mercaptopropionic acid, silanization, & polymer coating"
dx.doi.org/10.1021/nl0501…
doi.org/10.1088/0957-4…
(#12 and #9 were also about silver NPs)
But let's carry on.
dx.doi.org/10.1021/nl0523…
tandfonline.com/doi/full/10.34…
No information on the number of cells, vesicles, or nanoparticles analysed
ncbi.nlm.nih.gov/pmc/articles/P…

ncbi.nlm.nih.gov/pmc/articles/P…

dx.doi.org/10.1021/ja0572…