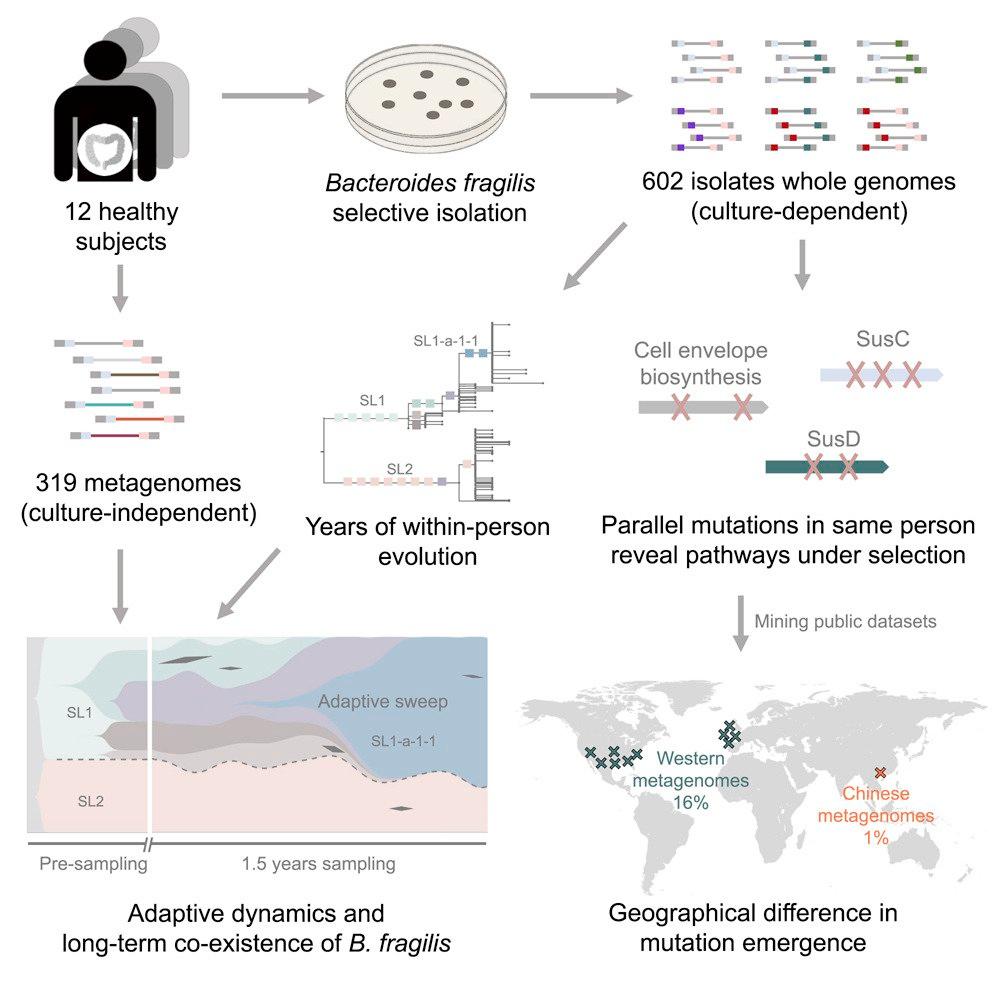
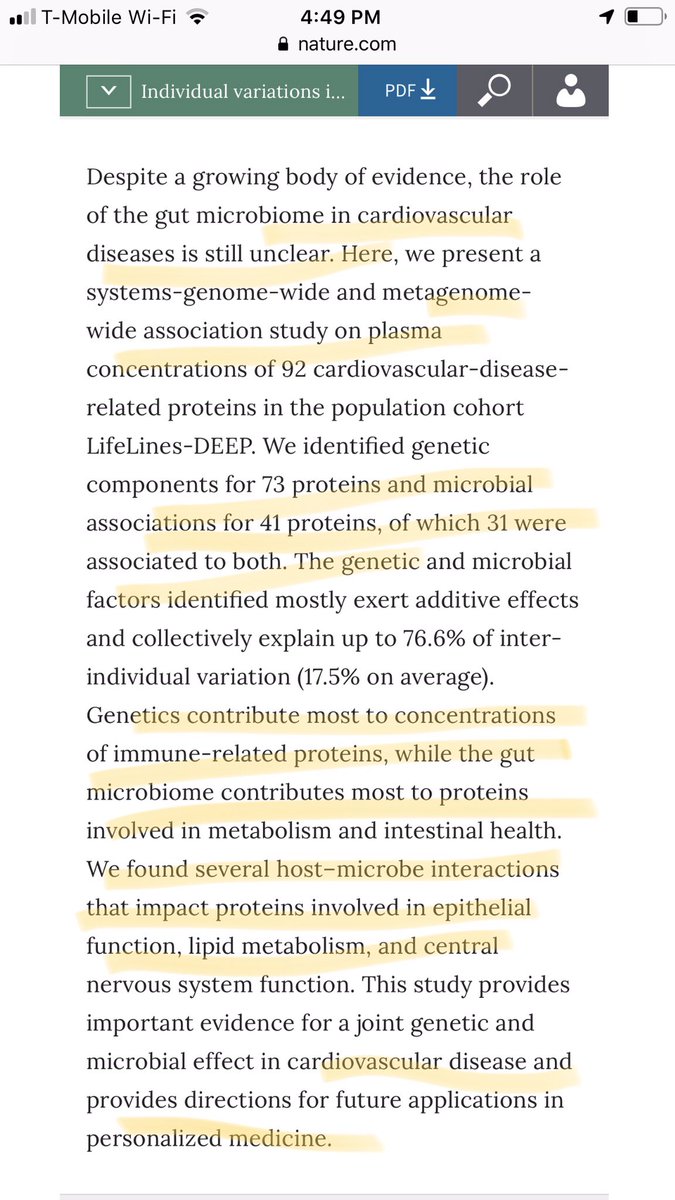
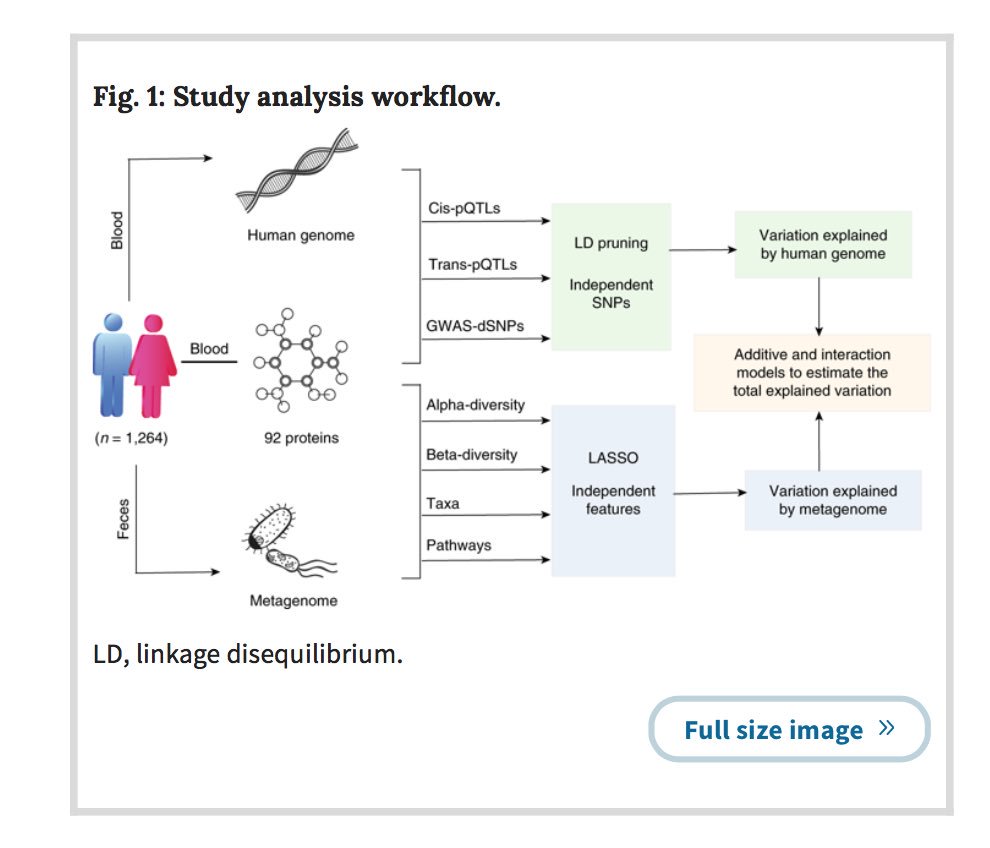
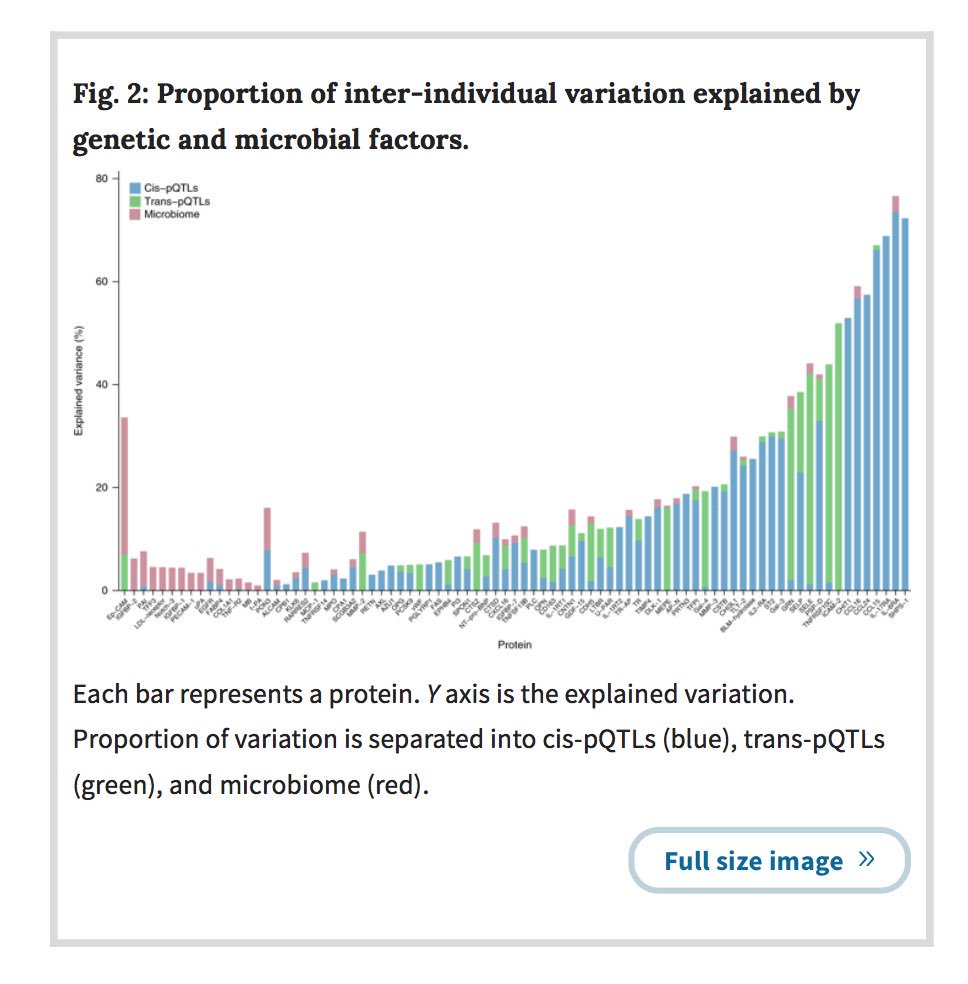
Your gut microbiome changes within you, even during health. What role do mutations play?
sciencedirect.com/science/articl…
Point mutations with strong adaptive effects have been seen in presence of antibiotics, in opportunistic infections (e.g. transition from soil to cystic fibrosis lung), and in lab experiments (e.g. lab strain to mice).
cell.com/cell-host-micr…
(lots of cool mice experiments)
journals.plos.org/plosbiology/ar…
(comparision between timepoints using metagenomics)