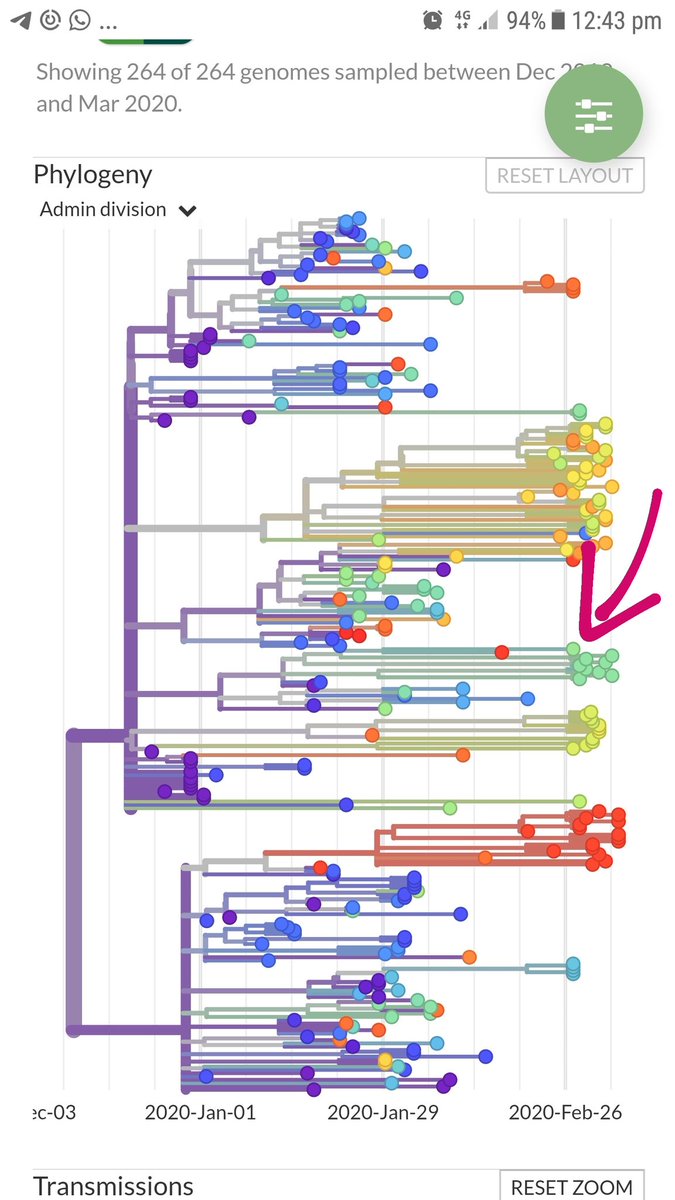
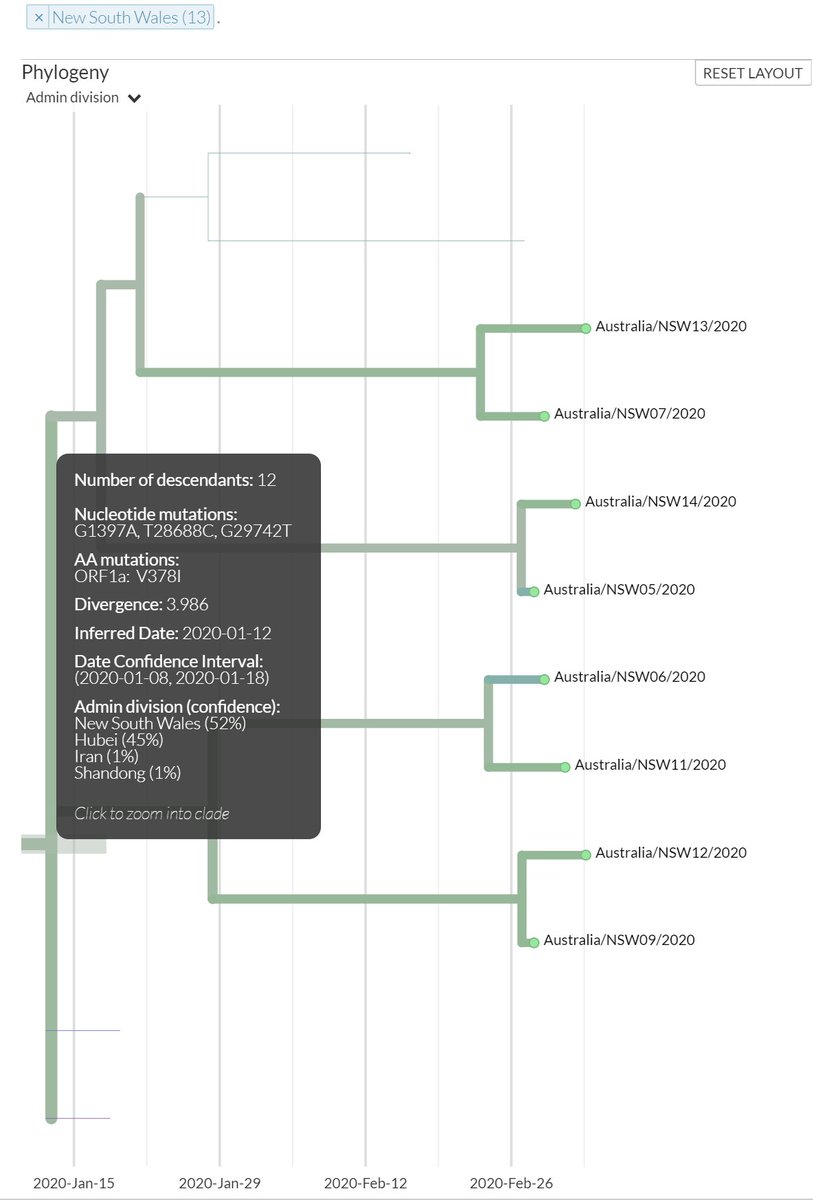
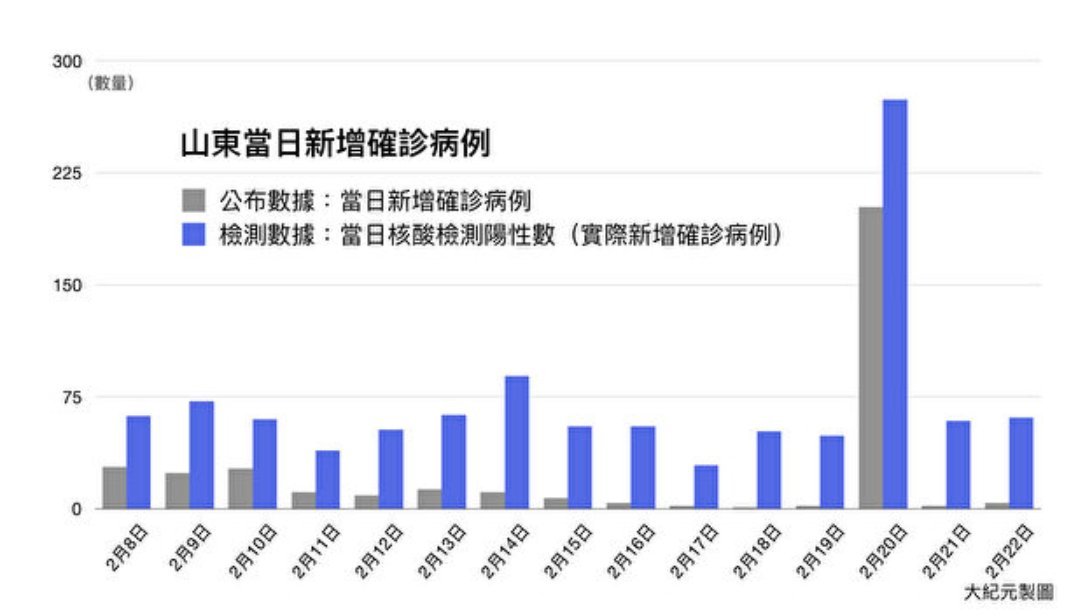
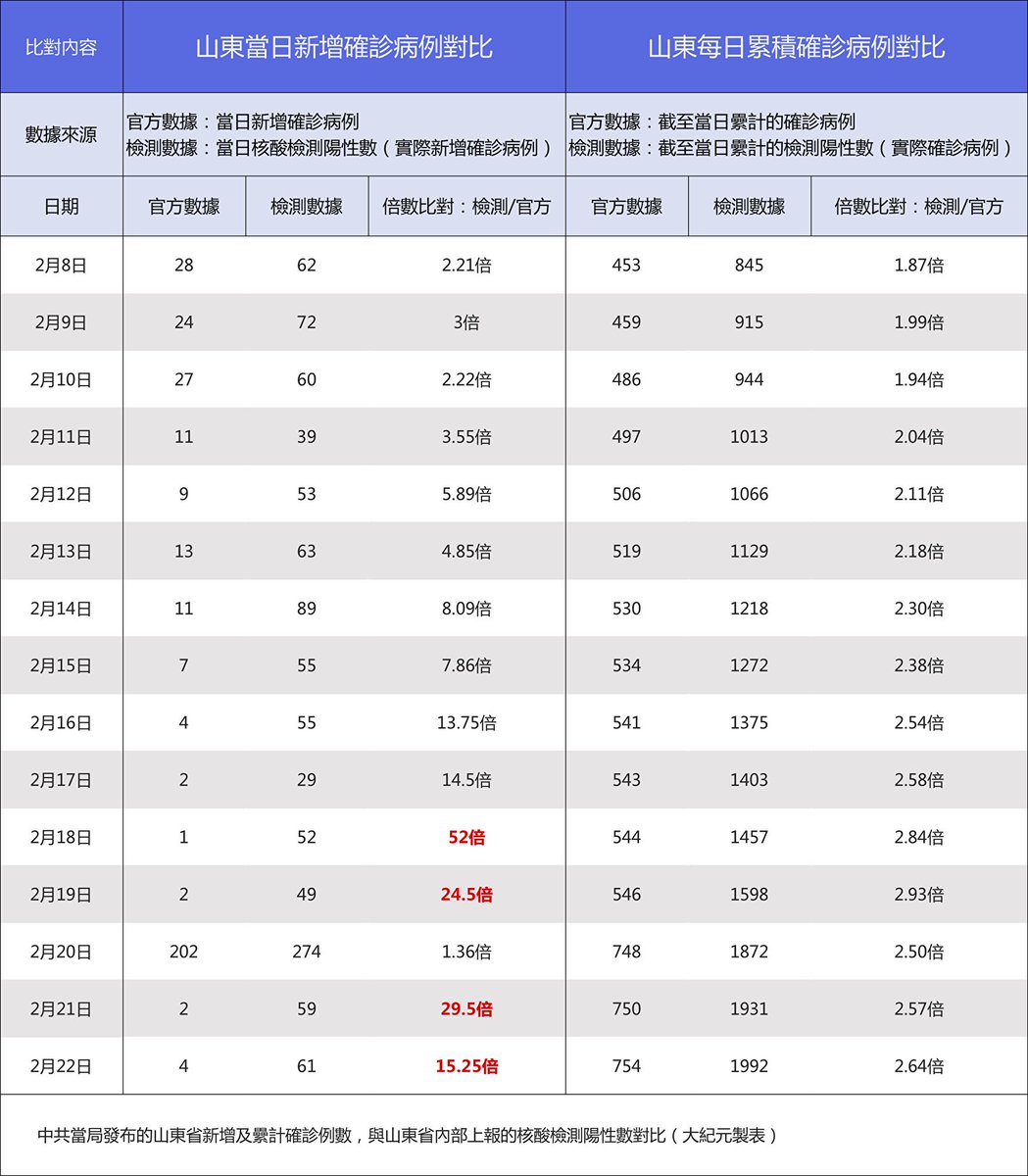
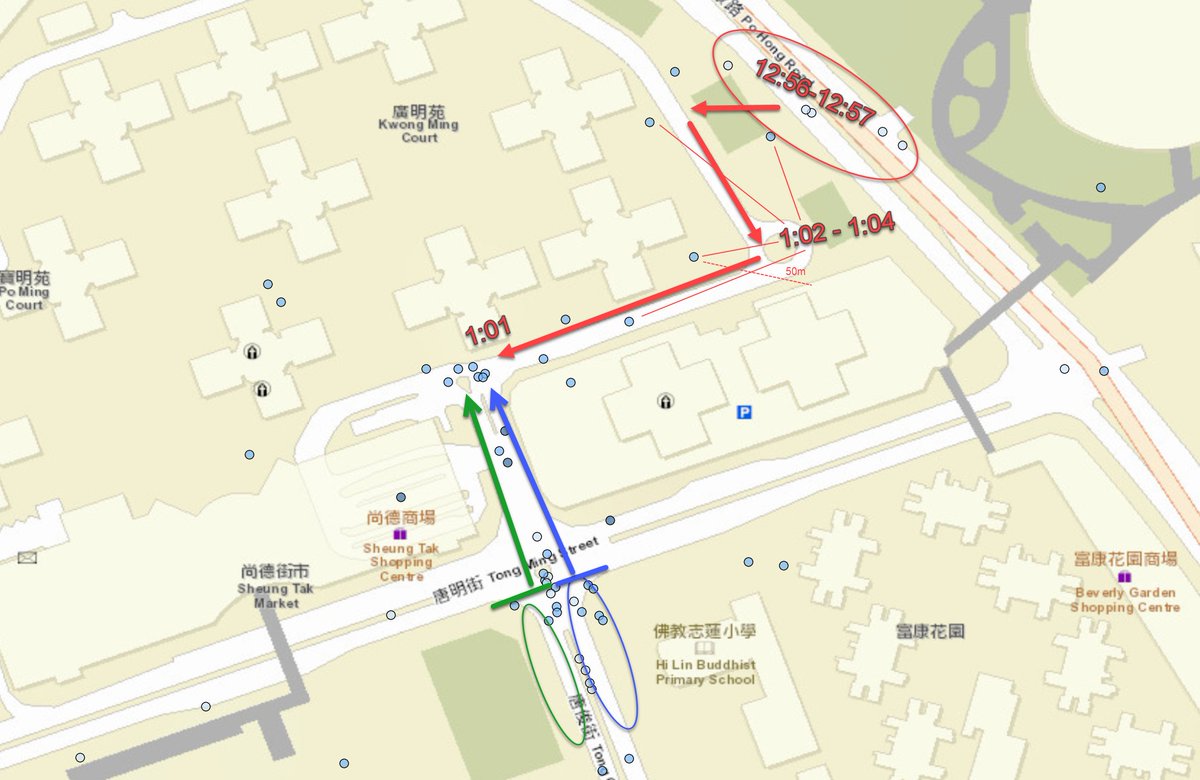
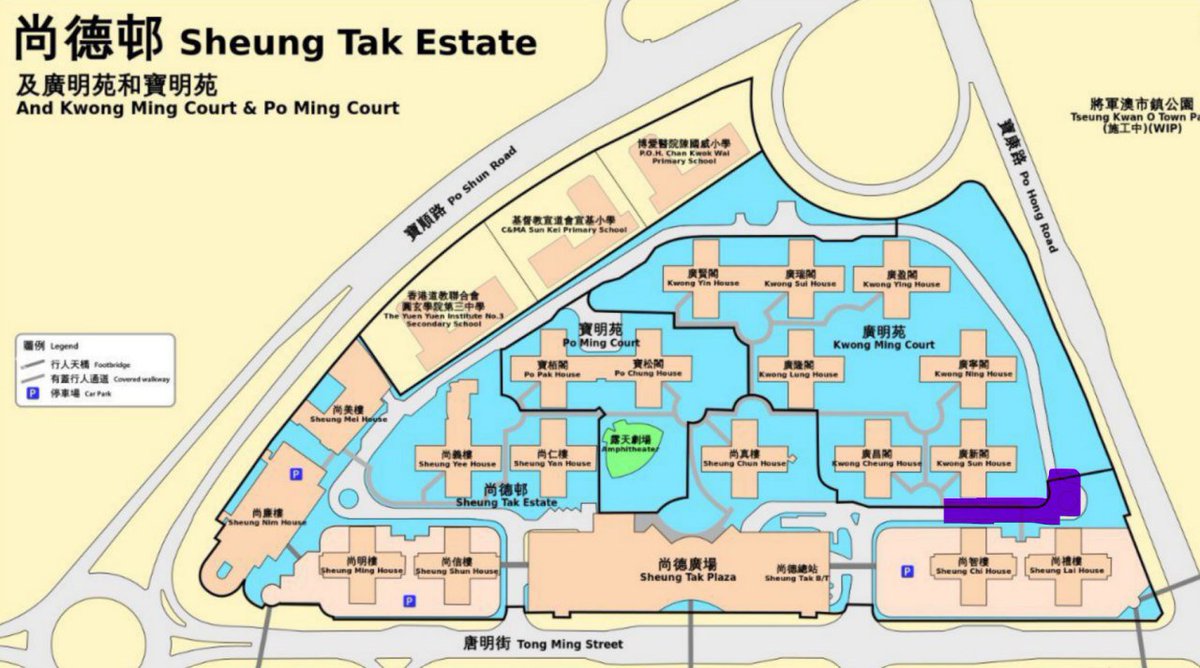
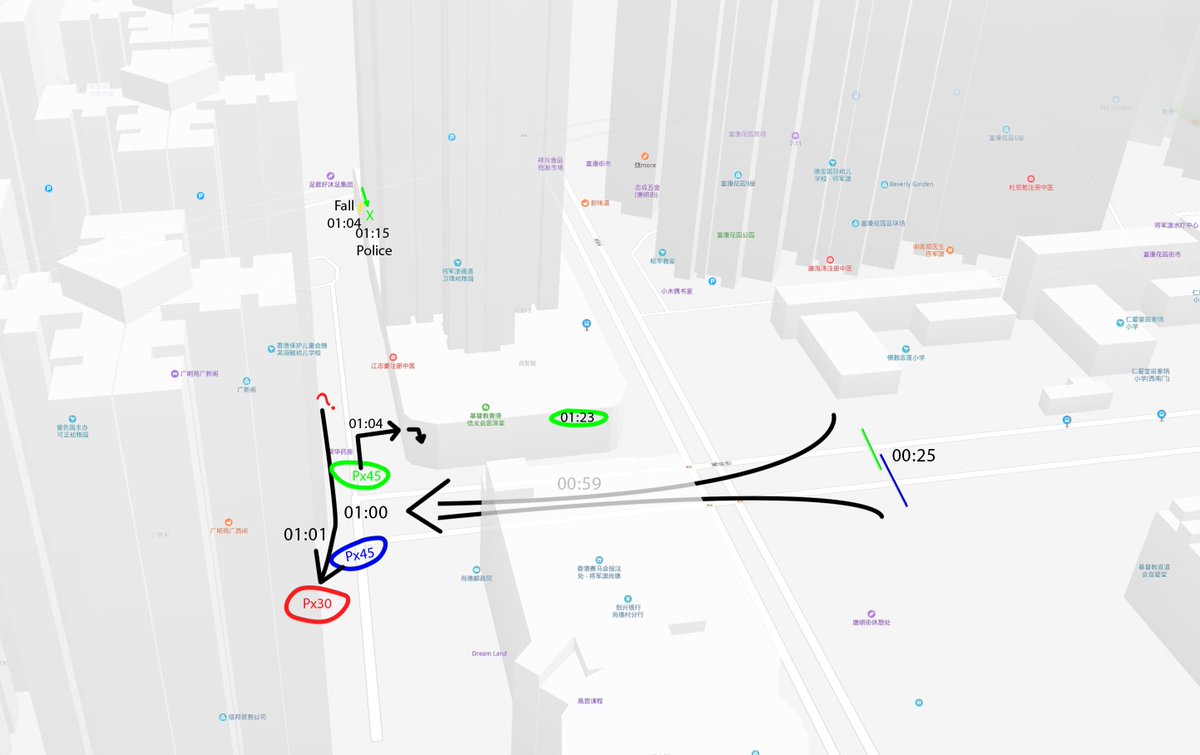
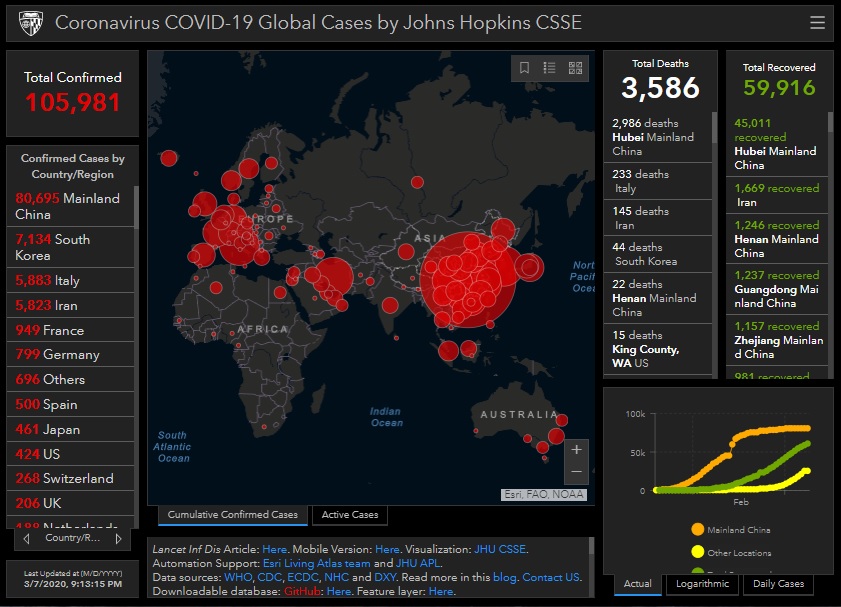
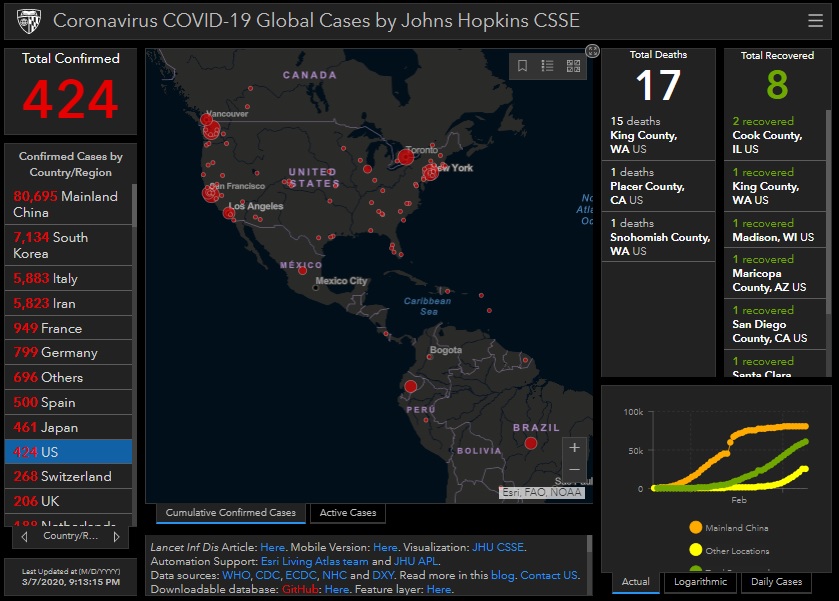
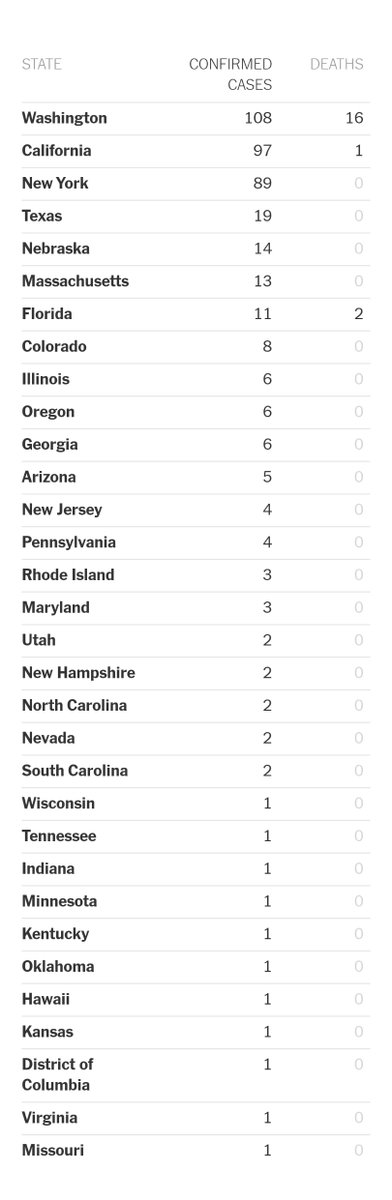
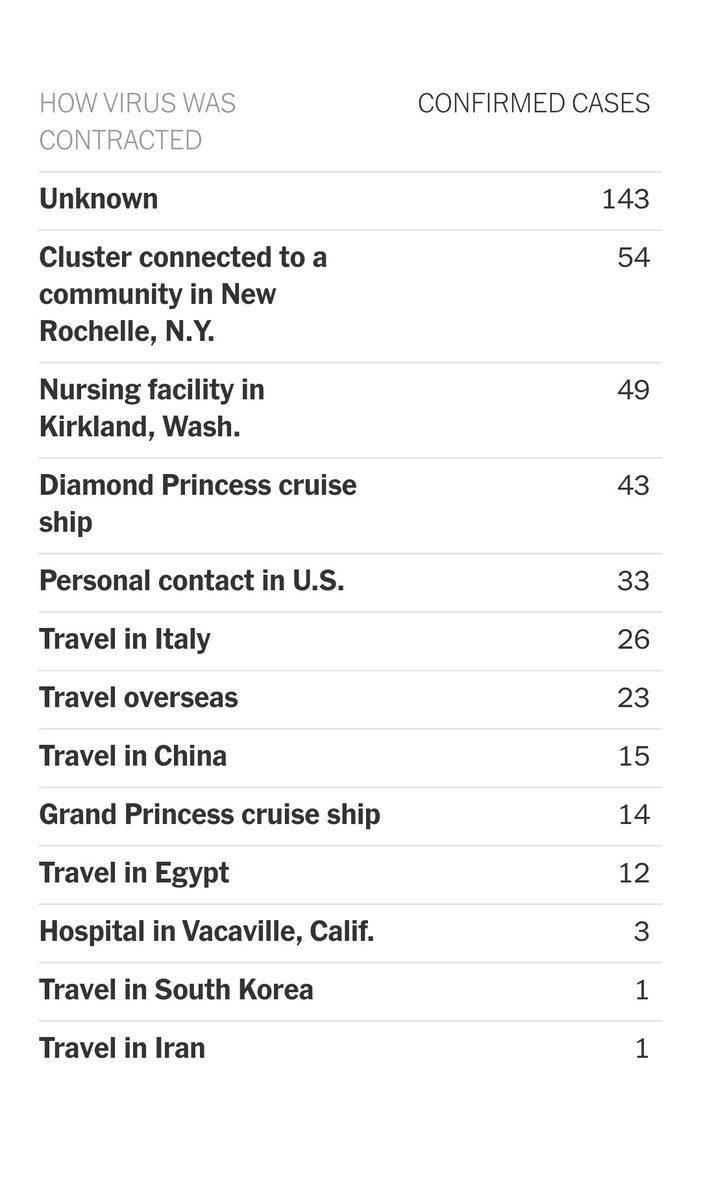
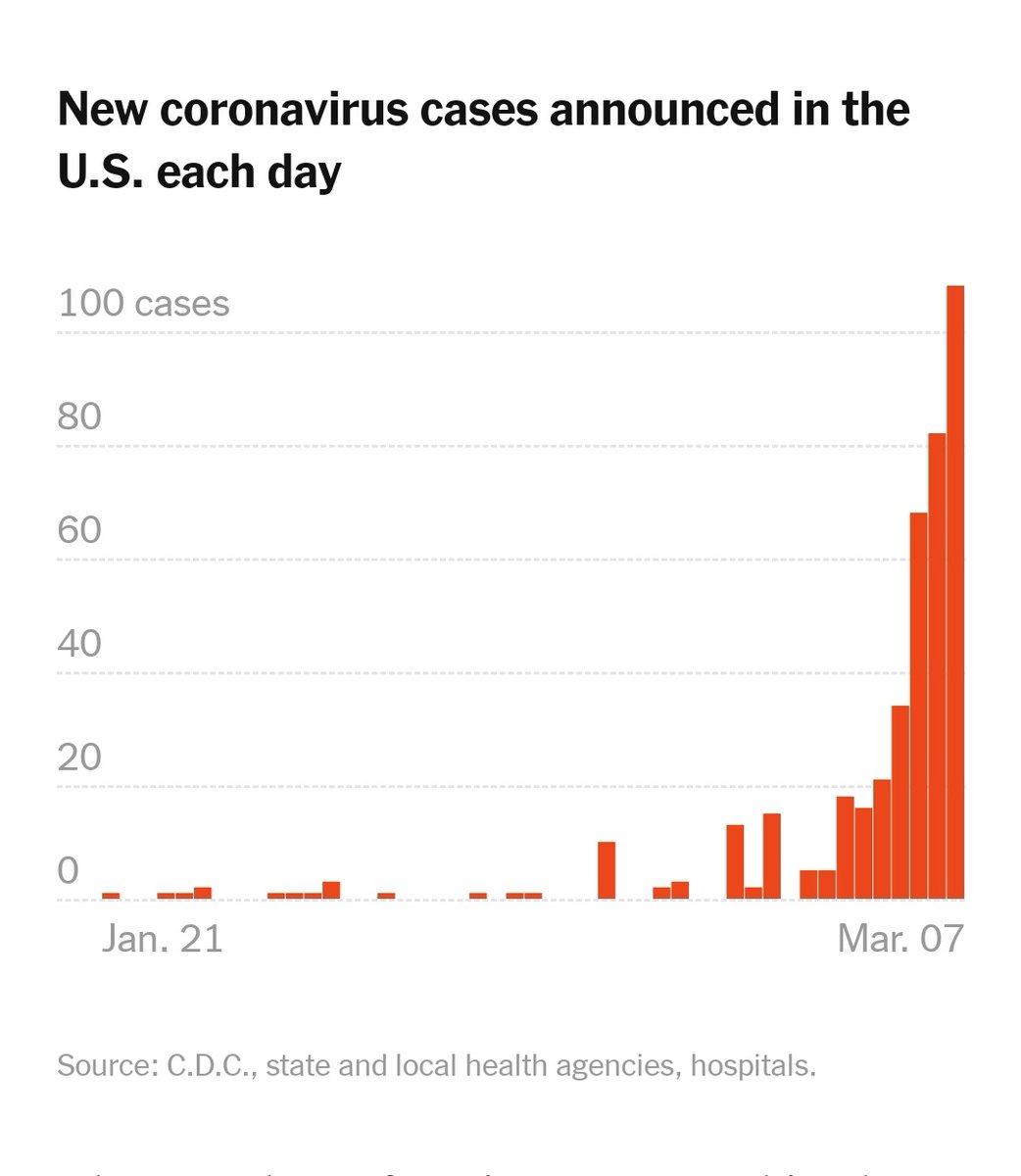
I have been looking at the genomics of Australia's #COVID2019 cases in recent days and I'm confident that there is significant and largely undetected community transmission of cases in Sydney, and there likely has been since Early Feb. Cases possibly total in the 100s
A THREAD








![Screenshot of a news headline: “[Op-ed] Why Vietnam has been the world’s number 1 country in dealing with coronavirus<br />
<br />
While other countries have dithered, Vietnam’s response to Covid-19 has been swift, sharp and effective”](https://pbs.twimg.com/media/ESjvKWiUUAEGprI.jpg)


